Search Count: 116
 |
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |
 |
Crystal Structure Of The Light-Driven Proton Pump Heimdallarchaeial Rhodopsin Heimdallr1
Organism: Candidatus heimdallarchaeota archaeon
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-02 Classification: PROTON TRANSPORT Ligands: RET, OLC, NO3 |
 |
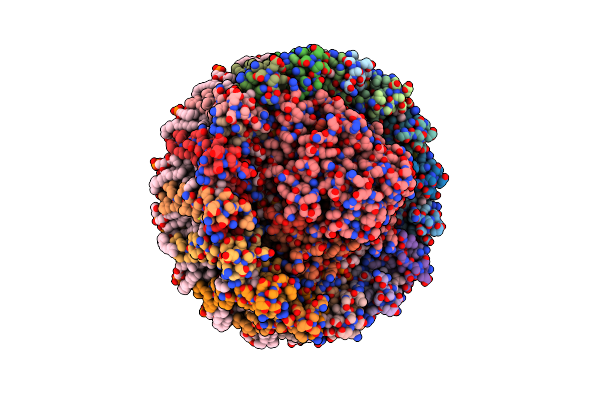
Photosynthetic Lh2-Lh1 Complex From The Purple Bacterium Halorhodospira Halophila
Organism: Halorhodospira halophila
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PHOTOSYNTHESIS Ligands: BCL, LMT, CRT, PGV |
 |
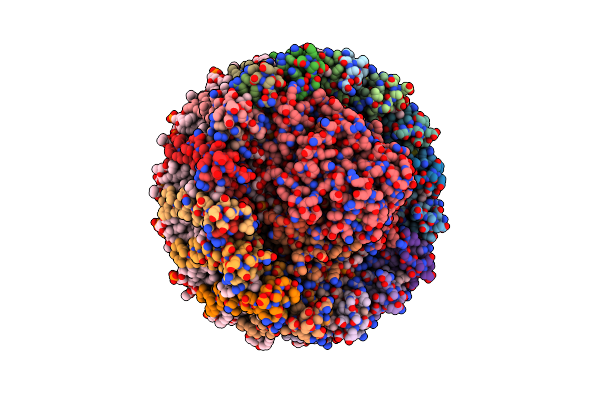
Photosynthetic Lh1-Rc-Hipip Complex From The Purple Bacterium Halorhodospira Halophila
Organism: Halorhodospira halophila
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: PHOTOSYNTHESIS Ligands: HEC, MG, LJQ, PLM, PGV, LMT, BCL, BPH, UQ8, FE, CDL, MQ8, CRT, SF4 |
 |
Photosynthetic Lh1-Rc Complex From The Purple Bacterium Halorhodospira Halophila
Organism: Halorhodospira halophila
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: PHOTOSYNTHESIS Ligands: HEC, MG, Z41, PLM, LMT, PGV, BCL, BPH, UQ8, CDL, FE, MQ8, CRT |
 |
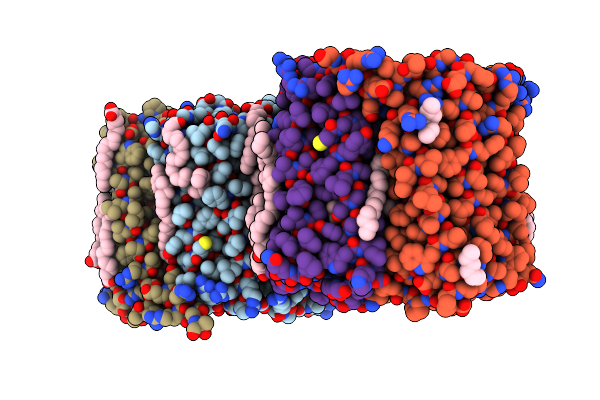
E. Coli 70S Ribosome Complexed With P. Putida Trnaile2 At The A-Site And P-Site
Organism: Escherichia coli, Pseudomonas putida nbrc 14164
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli, Escherichia coli bw25113, Pseudomonas putida nbrc 14164
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli, Escherichia coli bw25113, Pseudomonas putida nbrc 14164
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli, Escherichia coli bw25113, Pseudomonas putida nbrc 14164
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli, Escherichia coli bw25113, Pseudomonas putida nbrc 14164
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG |
 |
Organism: Salinarimonas soli
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN Ligands: LFA, CL, OLA, RET |
 |
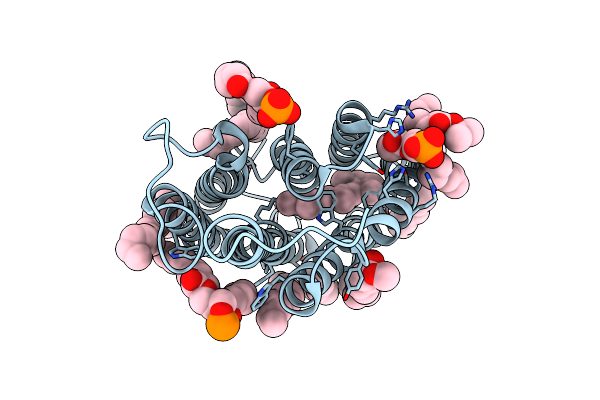
Neutron Structure Of Bacillus Thermoproteolyticus Ferredoxin At Room Temperature
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.45 Å, 1.60 Å Release Date: 2024-02-07 Classification: ELECTRON TRANSPORT Ligands: SF4 |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.56 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr2 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.53 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 H225F Mutant In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-05-17 Classification: LIPID BINDING PROTEIN Ligands: 3PE |
 |
Organism: Uncultured bdellovibrionales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: PROTON TRANSPORT Ligands: RET, K3I |

