Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Reducing-End Xylose-Releasing Exoxylanase In Gh30 From Talaromyces Cellulolyticus
Organism: Talaromyces pinophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-17 Classification: HYDROLASE Ligands: NAG, GOL, PG4, PEG |
 |
Crystal Structure Of Reducing-End Xylose-Releasing Exoxylanase In Gh30 From Talaromyces Cellulolyticus With Xylose
Organism: Talaromyces pinophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-17 Classification: HYDROLASE Ligands: NAG, GOL, XYP, CL |
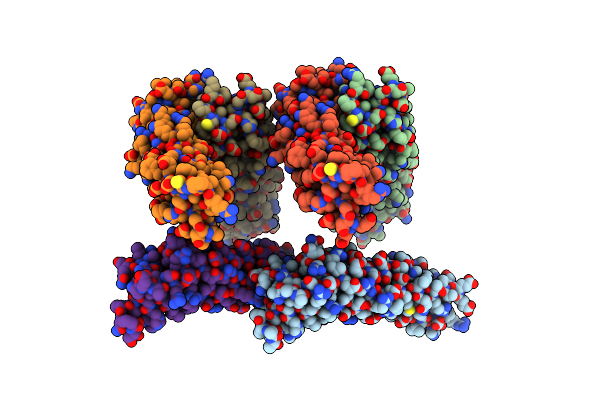 |
Cooperative Regulation Of Pbi1 And Mapks Controls Wrky45 Transcription Factor In Rice Immunity
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-04-06 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, CA |
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Daidzein- And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, ZF1, CA |
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Luteolin- And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, LU2, CA |
 |
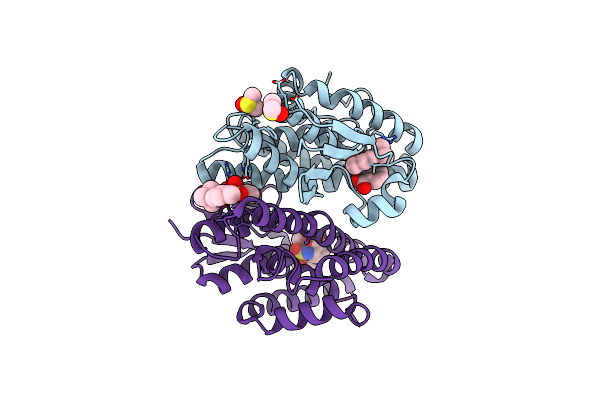
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Desmethylglycitein And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, 674, CA |
 |
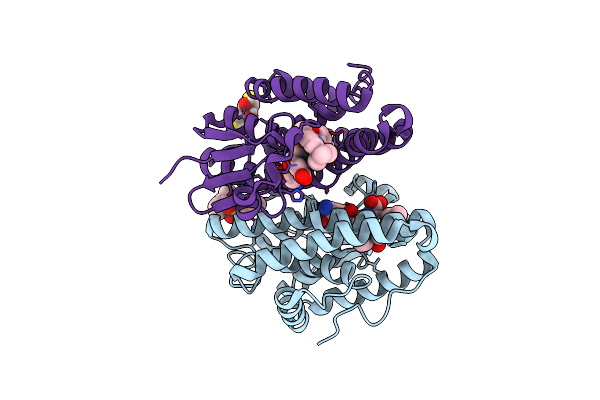
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp013-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: H1X, DMS, CL |
 |
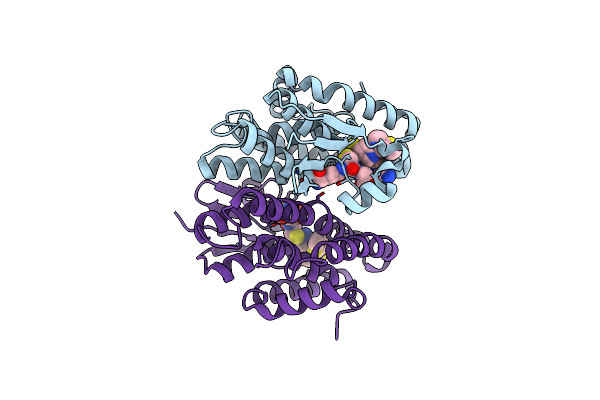
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp013-, And Gsh-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: H1X, GSH, DTT |
 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp015- And Gsh-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: GSH, H29 |
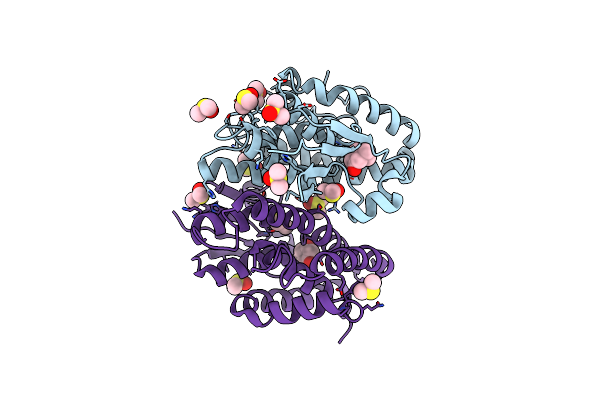 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Dimedone-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: DC1, DMS |
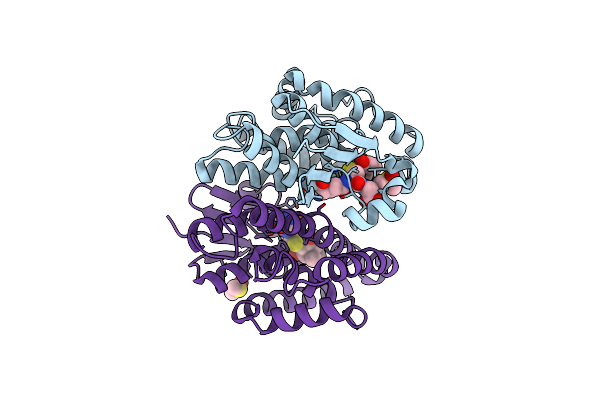 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Dimedone- And Glutathione-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: DC1, GSH, DMS |
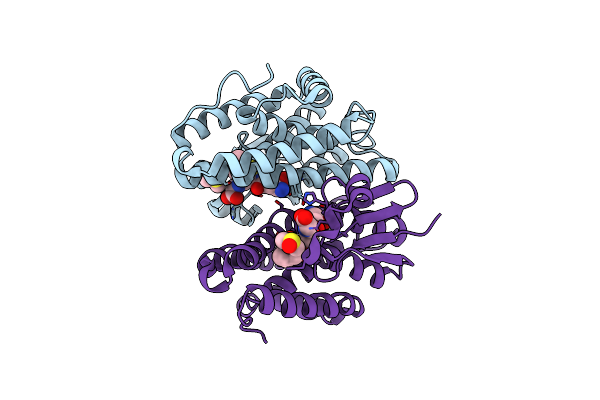 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Gs-Dimedone-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: DC1, GSH, DMS |
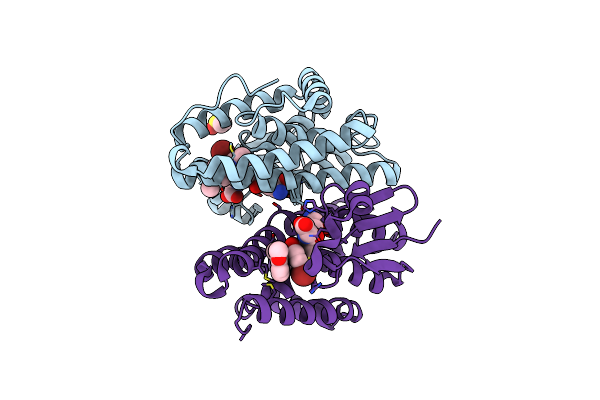 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp011-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: H2X, GSH, DMS |
 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp012- And Glutathione-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: H2C, GSH |
 |
Organism: Talaromyces cellulolyticus cf-2612
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-01-20 Classification: HYDROLASE Ligands: MAN, GOL, ACT |
 |
Crystal Structure Of Gh30 Xylanase B From Talaromyces Cellulolyticus Expressed By Pichia Pastoris
Organism: Saccharomyces uvarum, Talaromyces cellulolyticus cf-2612
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-06-17 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Crystal Structure Of Gh30 Xylanase B From Talaromyces Cellulolyticus Expressed By Pichia Pastoris In Complex With Aldotriuronic Acid
Organism: Saccharomyces uvarum, Talaromyces cellulolyticus cf-2612
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-06-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-11-13 Classification: OXIDOREDUCTASE Ligands: D8C, NAD, PO4 |
 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Apo-Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: TRS |