Search Count: 488
 |
Organism: Escherichia phage johannrwettstein
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage johannrwettstein
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage johannrwettstein
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage johannrwettstein
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRAL PROTEIN |
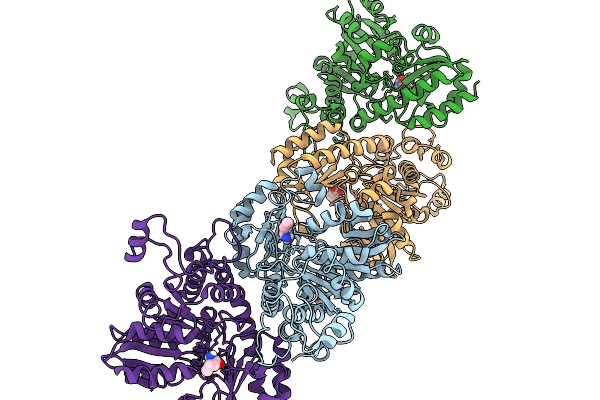 |
Crystal Structure Of Highly Stable Methionine Gamma-Lyase From Thermobrachium Celere In Complex With Plp And Norleucine
Organism: Thermobrachium celere
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: NLE, EDO |
 |
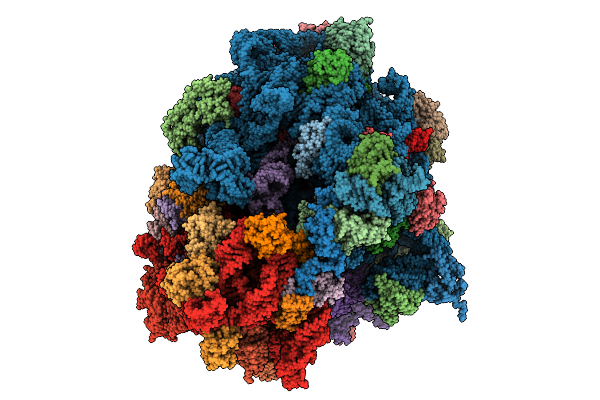
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 2 (H2-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, GDP, PO4 |
 |
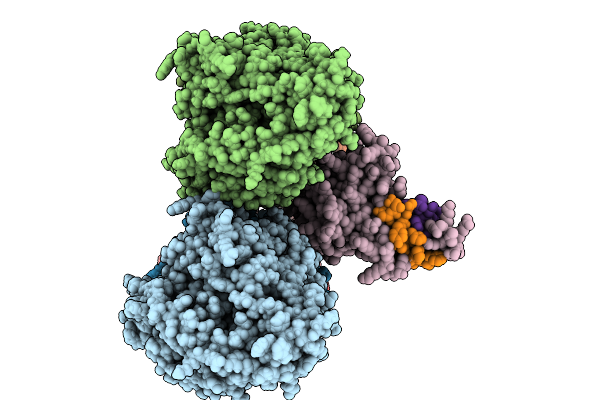
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 1 (H1-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, PO4, GDP |
 |
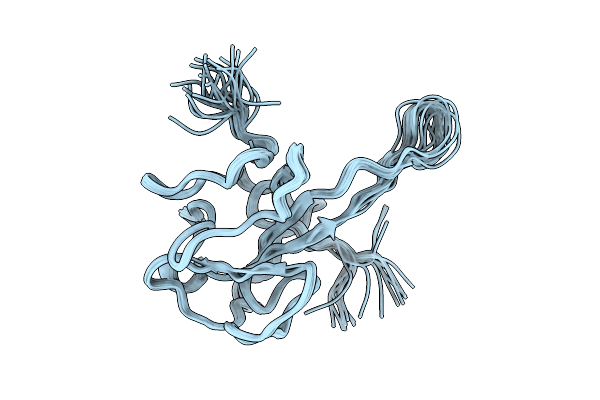
D. Melanogaster Augmin Tii N-Clamp (Gst-Fusion) Bound To A Microtubule, Well-Defined Subset Of Particles
Organism: Drosophila melanogaster, Aequorea victoria, Schistosoma japonicum, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: CELL CYCLE |
 |
Organism: Seneca valley virus usa/ssv-001
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRUS Ligands: CA |
 |
Seneca Valley Virus Altered Particle At Physiological Condition (A-Particle[P])
Organism: Seneca valley virus usa/ssv-001
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRUS Ligands: CA |
 |
Seneca Valley Virus Empty Rotated Particle At Acidic Condition (Er-Particle[C])
Organism: Seneca valley virus usa/ssv-001
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRUS Ligands: CA |
 |
Seneca Valley Virus Empty Rotated Particle At Physiological Condition (Er-Particle[P])
Organism: Seneca valley virus usa/ssv-001
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRUS Ligands: CA |
 |
The N-Terminal Domain (44-180) Of The Sars-Cov-2 Nucleocapsid Phosphoprotein Using An Automatic Assignment/Modeling Software
Organism: Severe acute respiratory syndrome coronavirus 2
Method: SOLUTION NMR Release Date: 2025-08-13 Classification: VIRAL PROTEIN |
 |
Structure Of In-Vivo Formed Alpha-Synuclein Fibrils Purified From A M83+/- Mouse Brain Injected With Recombinant 1B Fibrils
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN FIBRIL |
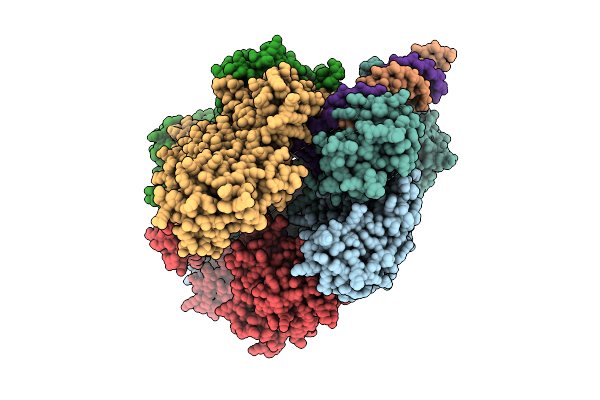 |
Structure Of The Bacteriophage Phikz Non-Virion Rna Polymerase Bound To An Analogue Of Its Promoter
Organism: Phikzvirus phikz
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: RNA BINDING PROTEIN Ligands: ZN |
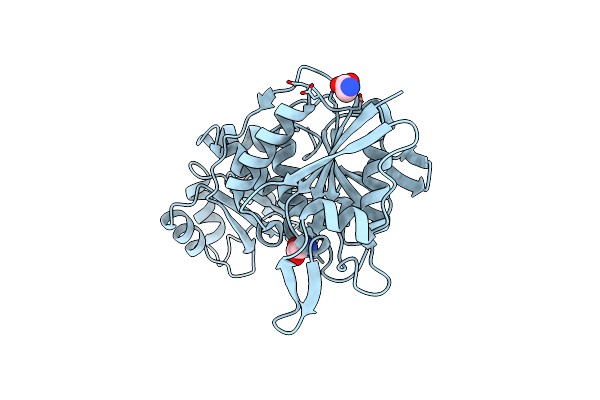 |
Organism: Thermococcus sibiricus
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: GLY |
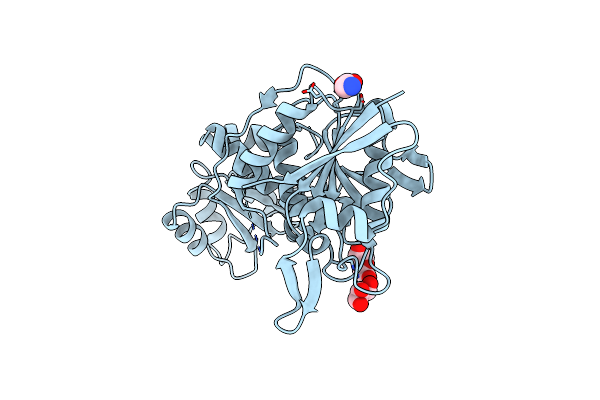 |
Crystal Structure Of L-Asparaginase From Thermococcus Sibiricus Double Mutation D54G/T56Q
Organism: Thermococcus sibiricus
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: PEG, GOL, GLY |
 |
Organism: Pectobacterium phage phite
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: VIRAL PROTEIN |
 |
Organism: Pectobacterium phage phite
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: VIRAL PROTEIN |
 |
Organism: Pectobacterium phage phite
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: VIRAL PROTEIN |

