Search Count: 45
 |
S-Methylcysteine Synthase (Bsas4) From Common Bean In Complex With Plp, Bez, And Gol
Organism: Phaseolus vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: BEZ, CL, GOL, NA |
 |
Crystal Structure Of Potassium-Independent L-Asparaginase From Phaseolus Vulgaris (Pvaiii, Pvaspg2)
Organism: Phaseolus vulgaris
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-05-14 Classification: HYDROLASE |
 |
Medicago Truncatula 5'-Profar Isomerase (Hisn3) D57N Mutant In Complex With Profar
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: GUO, NA, CL |
 |
Medicago Truncatula 5'-Profar Isomerase (Hisn3) D57N Mutant In Complex With Prfar
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: 2ER, MPD, NA, CL |
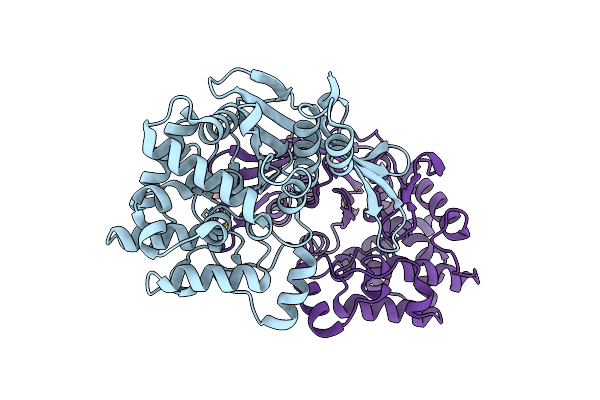 |
Crystal Structure Of Rhizobium Etli Constitutive L-Asparaginase Reaiv (Tetragonal Form R4Tp)
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: ZN, CL, EDO |
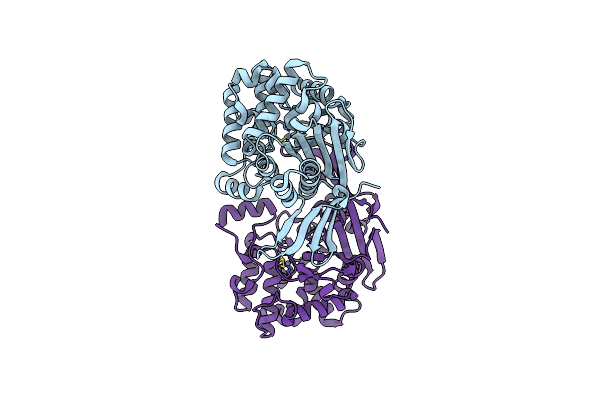 |
Crystal Structure Of Rhizobium Etli Constitutive L-Asparaginase Reaiv (Monoclinic Form R4Mc-2)
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: ZN, CL |
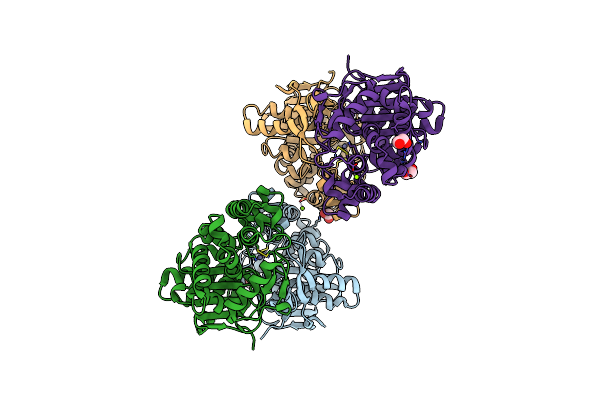 |
Crystal Structure Of Rhizobium Etli Constitutive L-Asparaginase Reaiv (Orthorombic Form R4Op-2)
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: ZN, CL, EDO, MG |
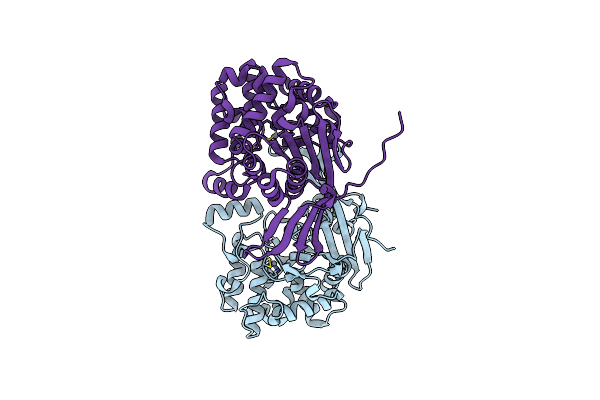
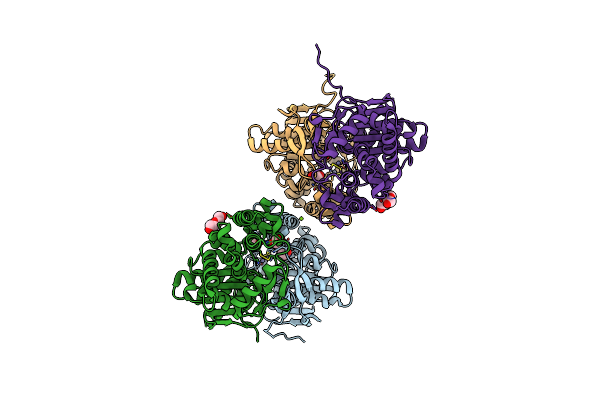 |
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: EDO, ZN, CL, PEG, MG, PGE |
 |
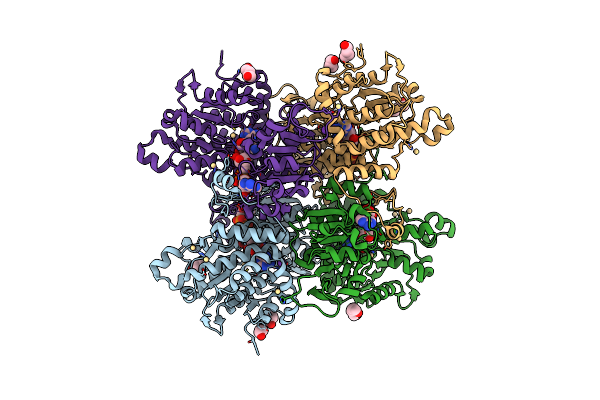
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: ZN, CL |
 |
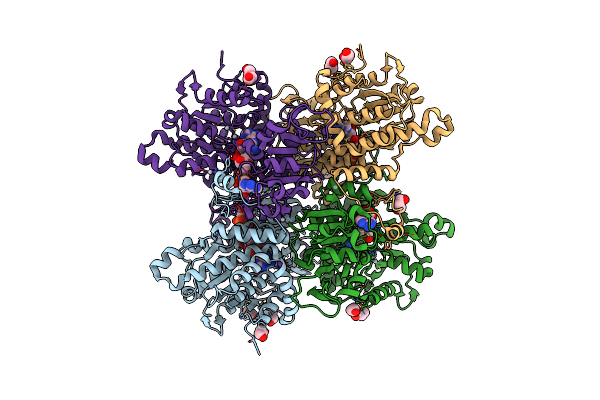
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Inhibited By Cd2+ Ions
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, BU1, PDO, K, CD, HEZ, CL |
 |
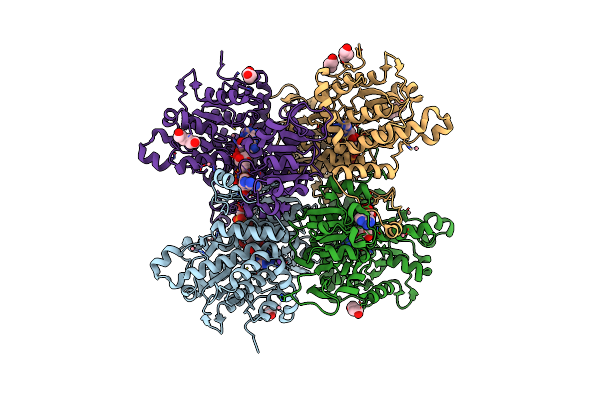
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Inhibited By Hg2+ Ions
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, BU1, PDO, K, HG, CL |
 |
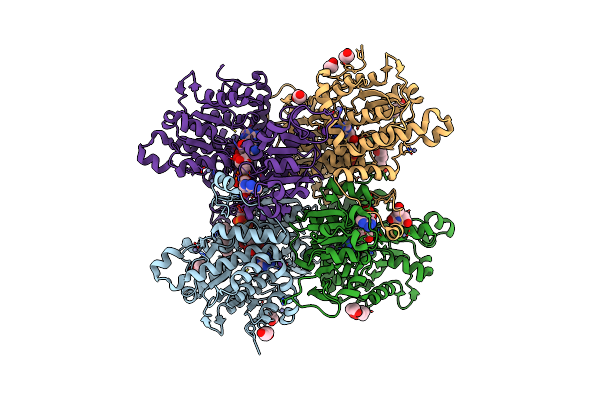
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Inhibited By Co2+ Ions.
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, PDO, K, CO, BU1, HEZ, CL |
 |
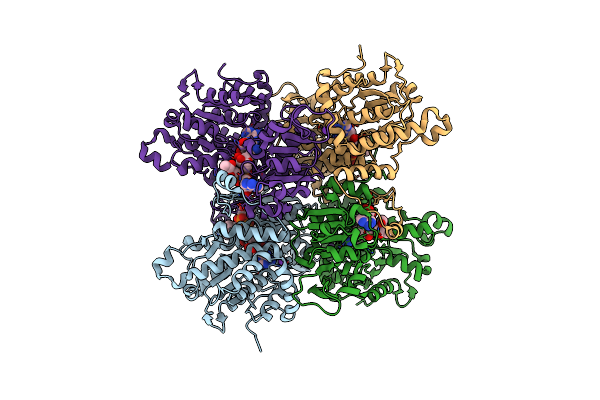
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Inhibited By Zn2+ Ions
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, BU1, PG4, K, ZN, BCN, HEZ, PDO, CL |
 |
Crystal Structure Of Pseudomonas Aeruginosa S-Adenosyl-L-Homocysteine Hydrolase Soaked With Cu+ Ions
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NAD, ADN, MPD, K, BR |
 |
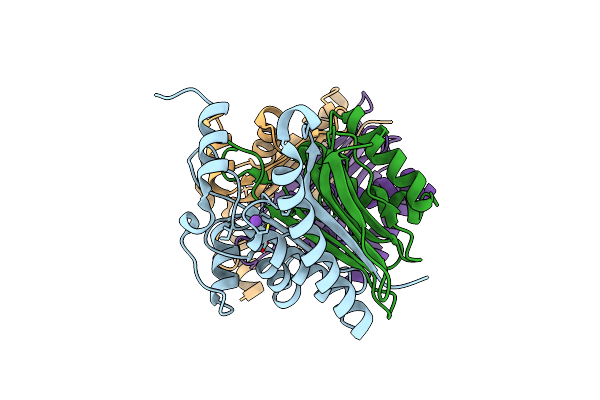
Structure Of E.Coli Class 2 L-Asparaginase Ecaiii, Mutant Rdm1-8 (G206Y, R207Q, D210P, S211T)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-13 Classification: HYDROLASE |
 |
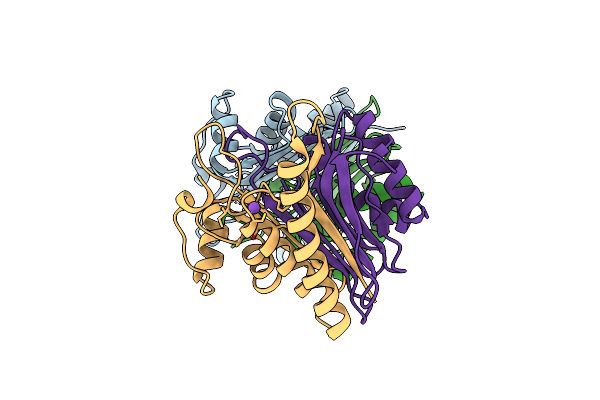
Structure Of E.Coli Class 2 L-Asparaginase Ecaiii, Mutant Rdm1-12 (G206C, R207T, D210A, S211A)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-13 Classification: HYDROLASE |
 |
Structure Of E.Coli Class 2 L-Asparaginase Ecaiii, Mutant Rdm1-3 (G206H, R207T, D210P, S211Q)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-07-13 Classification: HYDROLASE |
 |
Structure Of E.Coli Class 2 L-Asparaginase Ecaiii, Mutant Rdm1-37 (G206S, R207T, D210S)
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-13 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-07-13 Classification: HYDROLASE |
 |
Structure Of E.Coli Class 2 L-Asparaginase Ecaiii, Mutant Rdm1-18 (R207V, D210P, S211W)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-07-13 Classification: HYDROLASE |

