Search Count: 144
 |
Crystal Structure Of The Nucleotide-Binding Domain Of Candida Glabrata Hsp90
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: CHAPERONE |
 |
Structure Of Vibrio Vulnificus Martx Cysteine Protease Domain Lacking Beta-Flap
Organism: Vibrio vulnificus mo6-24/o
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-10 Classification: TOXIN |
 |
Organism: Vibrio vulnificus mo6-24/o
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-07-10 Classification: TOXIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-05-08 Classification: LIPID BINDING PROTEIN Ligands: IMD, GOL, NI |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN FIBRIL Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN FIBRIL |
 |
The Dbc1/Sirt1 Interaction Is Choreographed By Post-Translational Modification
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-27 Classification: GENE REGULATION |
 |
E.Coli Glyceraldehyde-3-Phosphate Dehydrogenase Structure Under Cryoprotect Condition Of Ammonium Sulfate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-02-14 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Crystal Structure Of The Ligand-Binding Domain Of C. Glabrata Upc2 In Complex With Ergosterol
Organism: [candida] glabrata cbs 138
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2022-09-07 Classification: TRANSCRIPTION Ligands: ERG |
 |
Organism: [candida] glabrata cbs 138
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-09-07 Classification: PROTEIN TRANSPORT |
 |
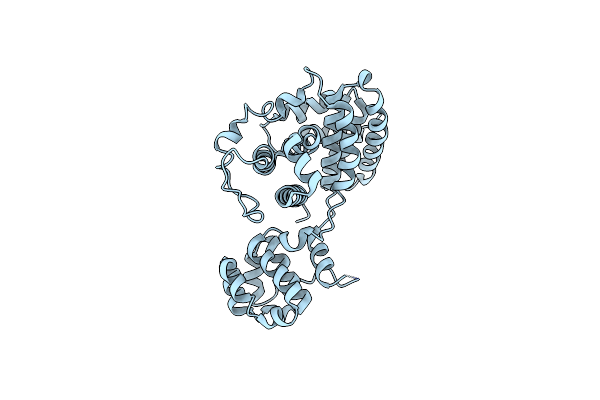
Organism: [candida] glabrata cbs 138
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-09-07 Classification: PROTEIN TRANSPORT |
 |
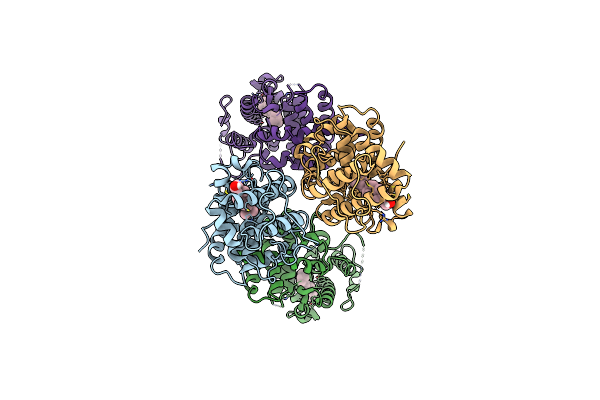
Structure Of The Ligand-Binding Domain Of S. Cerevisiae Upc2 In Fusion With T4 Lysozyme
Organism: Saccharomyces cerevisiae s288c, Escherichia virus t4
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2022-09-07 Classification: TRANSCRIPTION |
 |
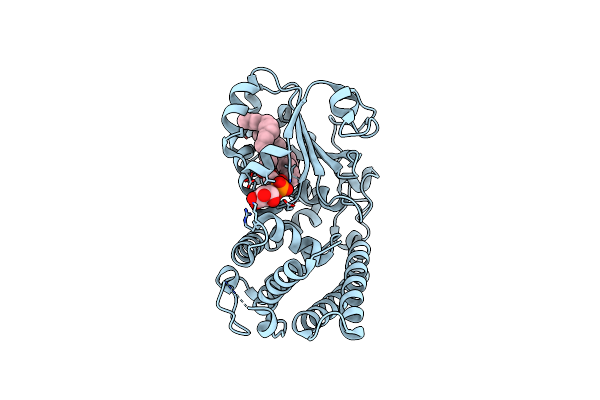
Crystal Structure Of The Ligand-Binding Domain Of L. Thermotolerans Upc2 In Complex With Ergosterol
Organism: Lachancea thermotolerans cbs 6340
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2022-08-24 Classification: TRANSCRIPTION Ligands: ERG |
 |
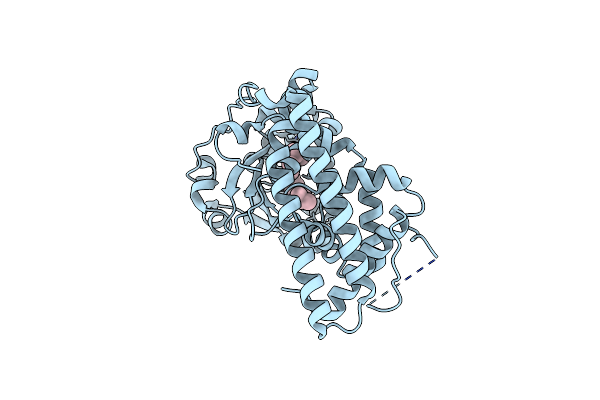
Crystal Structure Of Saccharomyces Cerevisiae Sfh2 Complexed With Phosphatidylinositol
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-07-13 Classification: LIPID TRANSPORT Ligands: B7N |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2022-07-13 Classification: LIPID TRANSPORT Ligands: SQL |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-13 Classification: LIPID TRANSPORT |
 |
Crystal Structure Of Saccharomyces Cerevisiae Sfh2 Complexed With Phosphatidylinositol In An Open Conformation
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-07-13 Classification: LIPID TRANSPORT Ligands: B7N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-23 Classification: LIPID TRANSPORT Ligands: MG, GTP |
 |
Organism: Daphnia magna
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-02-16 Classification: TRANSFERASE Ligands: PO4, NO3 |
 |
X-Ray Crystal Structure Of Visual Arrestin Complexed With Inositol Hexaphosphate
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-10-27 Classification: SIGNALING PROTEIN Ligands: IHP |

