Search Count: 265
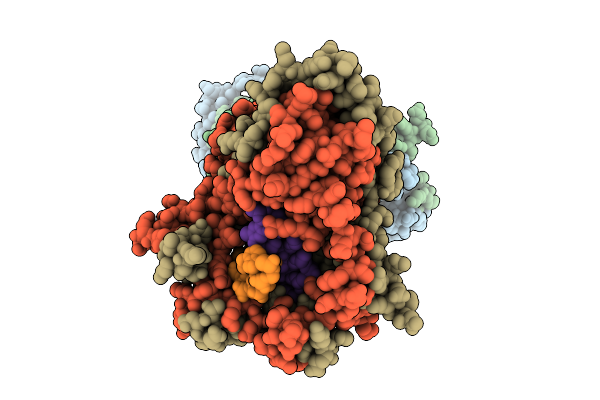 |
Organism: Bacillus subtilis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DNA BINDING PROTEIN |
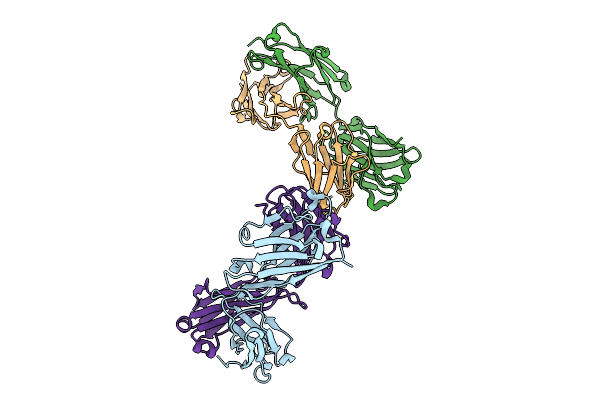 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
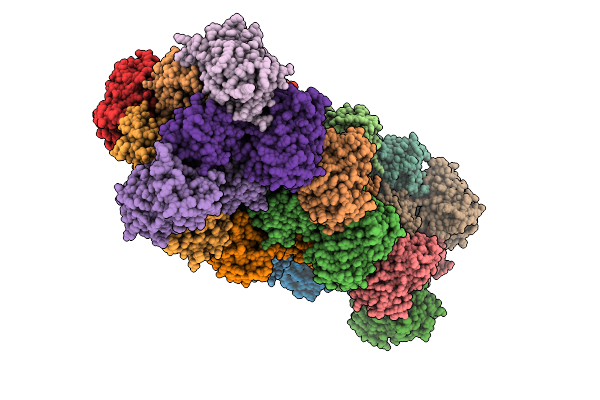 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: FLUORESCENT PROTEIN Ligands: NO3, SO4 |
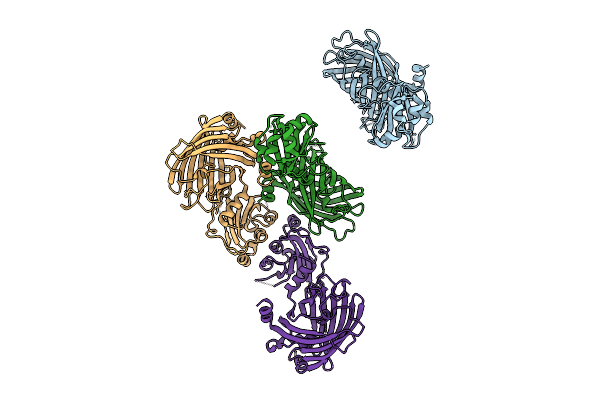 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: FLUORESCENT PROTEIN |
 |
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSFERASE |
 |
Crystal Structure Of Aprg Complexed With A Two-Carbon Amino Sugar Fragment (Acetamidoacetaldehyde)
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSFERASE Ligands: A1AU8, MG, ACT |
 |
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSFERASE Ligands: A1AVA |
 |
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSFERASE Ligands: A1AU8, ACT, A1AVJ |
 |
Organism: Gammaproteobacteria
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: URE |
 |
Organism: Gammaproteobacteria, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: DNA BINDING PROTEIN Ligands: MN, DCP |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: TRS, CU |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: TRS, CU |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, TYR |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, I3J |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PG4, A1A46, PO4, EDO, PG0, PG6 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: R75 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: A1AIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: A1A45 |
 |
Organism: Kitasatospora setae
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: BIOSYNTHETIC PROTEIN |

