Search Count: 1,940
 |
Organism: Faecalibacterium prausnitzii l2-6
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
 |
Organism: Faecalibacterium prausnitzii l2-6
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Glucocorticoid Receptor Ligand Binding Domain In Complex With Dexamethasone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN Ligands: DEX, CAC, EPE, GOL, SO4, CL, PG4, IMD, PEG, PGE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: CA, YZY, Y01, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: PTY, POV, Y01, ZN, CA, YZY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: PTY, POV, Y01, ZN, CA, YZY |
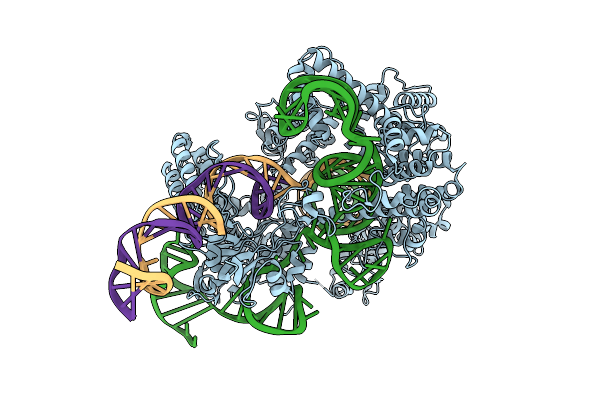 |
Crystal Structure Of Spcas9 Ternary Complex, Amino Acids (1242-1263) Replaced With Gly-Ser Linker
Organism: Streptococcus pyogenes, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN |
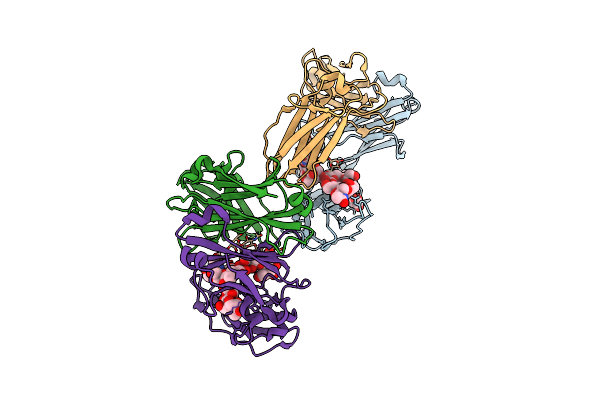 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: GAL |
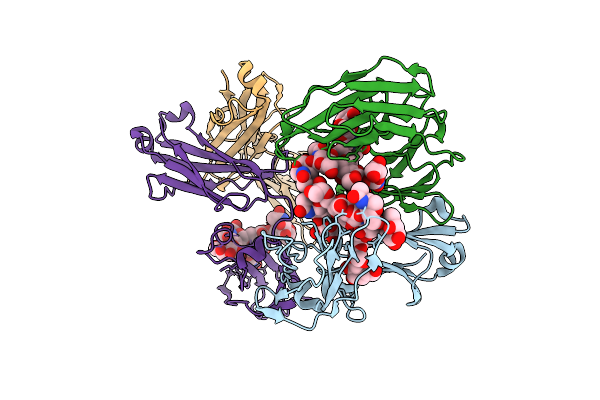 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: FUL |
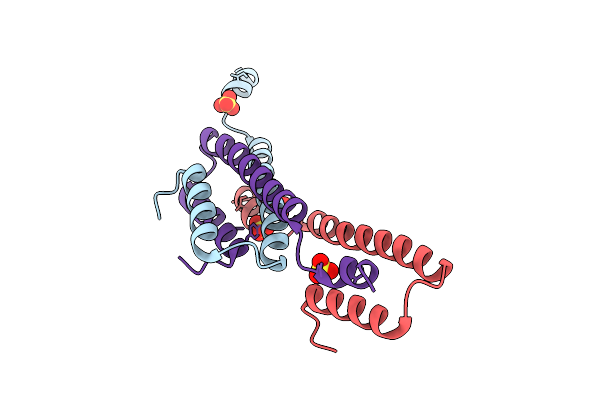 |
Crystal Structure Of The Semet-Derived C-Terminal Of Viral Responsive Protein 15 (Pmvrp15) From Black Tiger Shrimp Penaeus Monodon
Organism: Penaeus monodon
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Crystal Structure Of The C-Terminal Of Viral Responsive Protein 15 (Pmvrp15) From Black Tiger Shrimp Penaeus Monodon
Organism: Penaeus monodon
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Iron-Bound Human Ado C18S/C239S Variant Soaked In Hydralazine At 2.39 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FE2, HLZ, GOL |
 |
Crystal Structure Of Cobalt-Bound Human Ado C18S/C239S Variant In Complex With Hydralazine At 1.89 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: OXIDOREDUCTASE Ligands: CO, HLZ, GOL, SO4 |
 |
Crystal Structure Of The Mu2 Subunit Of The Clathrin-Adaptor Protein 2 (Ap2) Bound To Hpv16 E7(Residues 22-39)
Organism: Rattus norvegicus, Human papillomavirus 16
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ENDOCYTOSIS |
 |
Crystal Structure Of The Mu2 Subunit Of The Clathrin-Adaptor Protein 2 (Ap2) Bound To Hpv16 E7(Residues 22-32; S31E And S32E)
Organism: Rattus norvegicus, Human papillomavirus 16
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ENDOCYTOSIS |
 |
Crystal Structure Of The Mu2 Subunit Of The Clathrin-Adaptor Protein 2 (Ap2) Bound To Hpv16 E7(Residues 22-39; S31E And S32E)
Organism: Rattus norvegicus, Human papillomavirus 16
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ENDOCYTOSIS |
 |
Organism: Fasciola hepatica
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: CELL ADHESION |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBOTOR Ligands: A1L7M |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |

