Search Count: 208
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: A1BH7 |
 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001897 (Compound 1)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 5VA, CU |
 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001877 (Compound 39)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 648, CU |
 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001886 (Compound 38)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 5VB, CU, GOL |
 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001896 (Compound 72)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 62J, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: CU, 010 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: IMD |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: BYZ, PGE |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: PKN, CU |
 |
Crystal Structure Of Ecdsba In A Complex With (1-Methyl-1H-Pyrazol-5-Yl)Methanamine
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q2I, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q2O, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q3F, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: URE, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: DMS, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: GOL, DMS, CU |
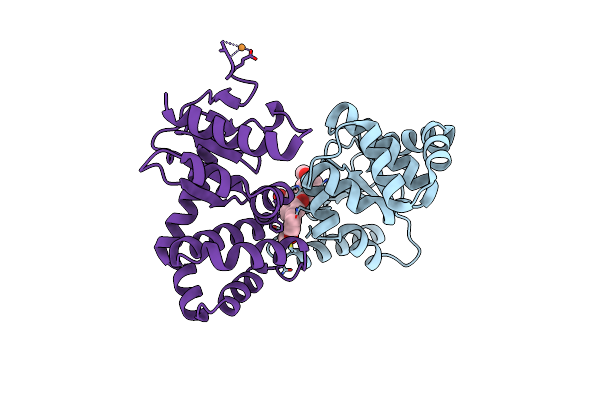 |
Crystal Structure Of E.Coli Dsba In Complex With 2-(2,6-Bis(3-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VCY, CU |
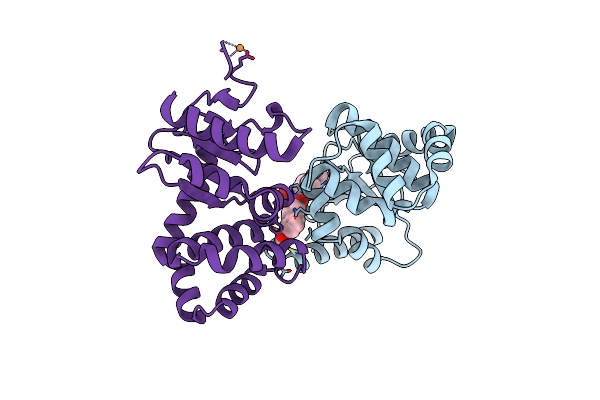 |
Crystal Structure Of E.Coli Dsba In Complex With 2-(6-(3-Methoxyphenyl)-2-(4-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VE7, CU |
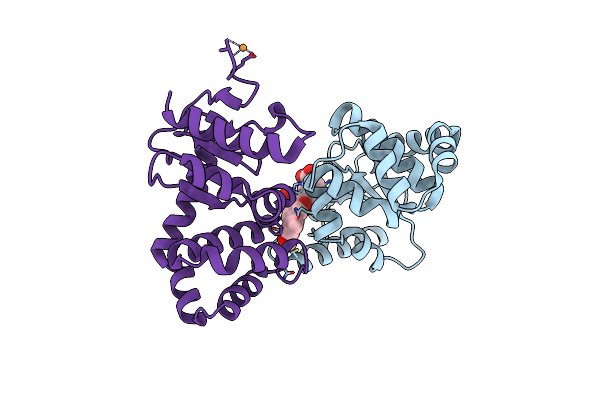 |
Crystal Structure Of E.Coli Dsba In Complex With 3-(3-(Carboxymethyl)-6-(3-Methoxyphenyl)Benzofuran-2-Yl)Benzoic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VED, CU |
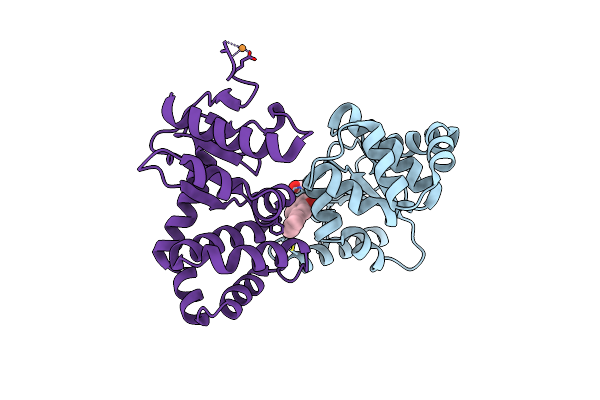 |
Crystal Structure Of Ecdsba In A Complex With 2-(6-Phenylbenzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: XPV, CU |
 |
Crystal Structure Of Ecdsba In A Complex With 2-(6-(3-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: XQ1, CU |

