Search Count: 20
 |
Cryoem Structure Of The F Plasmid Relaxosome In Its Pre-Initiation State, Derived From The Ds-27_+143-R Locally-Refined Map 3.76 A
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R Locally-Refined 3.45 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Truncated Trai1-863 In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R_Deltaah+Ctd Locally-Refined 3.42 A Map
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Without Accessory Protein Tram. Derived From The Ss-27_+8Ds+9_+143-R_Deltatram Locally-Refined 2.94 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Orit Dna Ss-27_+3Ds+4_+143 And Trai Its Te Mode, Derived From Ss-27_+3Ds+4_+143-R Locally-Refined 3.68 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Orit Dna Ss-27_-3Ds-2_+143 And Trai Its Te Mode, Derived From Ss-27_-3Ds-2_+143-R Locally-Refined 3.42 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of The Bacterial Replication Origin Opening Basal Unwinding System
Organism: Bacillus subtilis, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: REPLICATION Ligands: MG, ATP |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: ANP, K, MG |
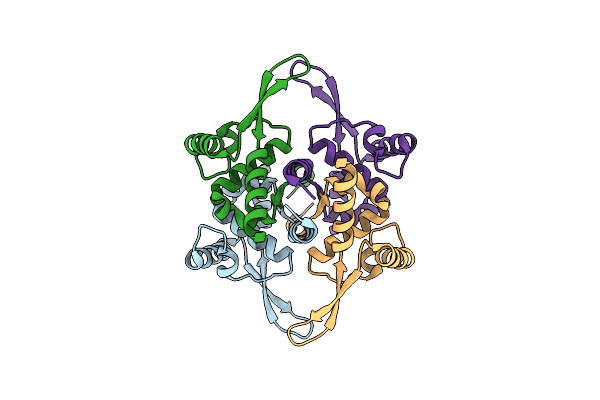 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: DNA BINDING PROTEIN |
 |
Structure Of The Outer-Membrane Core Complex (Outer Ring) From A Conjugative Type Iv Secretion System
Organism: Salmonella enterica, Salmonella enterica subsp. salamae serovar 58:l,z13,z28:z6
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: MEMBRANE PROTEIN |
 |
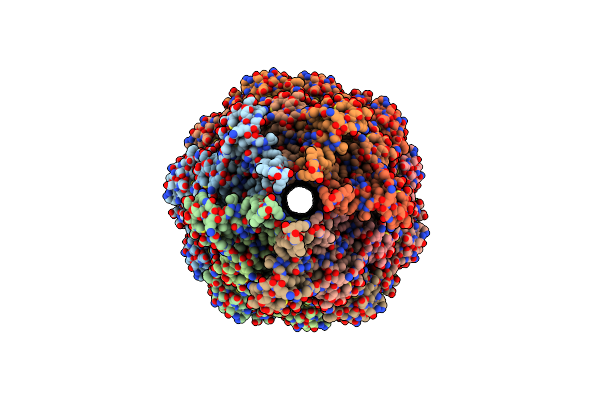
Structure Of The Outer-Membrane Core Complex (Inner Ring) From A Conjugative Type Iv Secretion System
Organism: Salmonella enterica
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: MEMBRANE PROTEIN |
 |
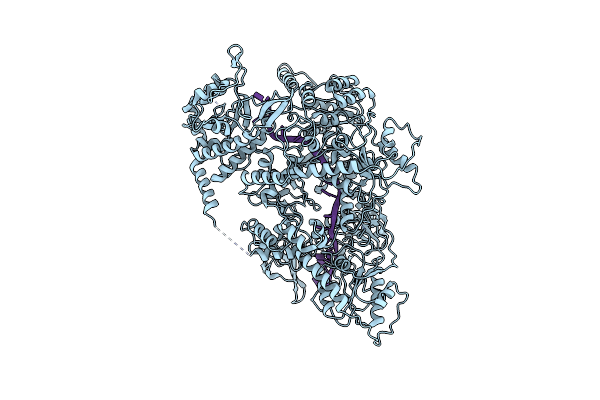
Cryo-Em Structure Of Enteropathogenic Escherichia Coli Type Iii Secretion System Espa Filament
Organism: Escherichia coli o127:h6 (strain e2348/69 / epec)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-23 Classification: PROTEIN FIBRIL |
 |
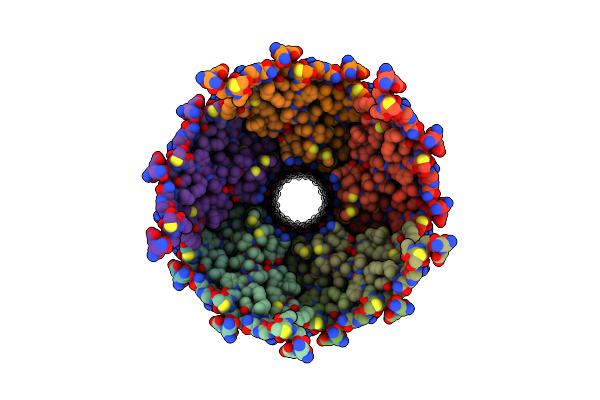
Cryo Em Structure Of The Conjugative Relaxase Trai Of The F/R1 Plasmid System
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2017-05-03 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2016-11-02 Classification: PROTEIN FIBRIL Ligands: 6V6 |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2016-11-02 Classification: PROTEIN FIBRIL Ligands: 6V6 |
 |
Organism: Salmonella enterica subsp. enterica serovar typhi
Method: ELECTRON MICROSCOPY Release Date: 2016-09-28 Classification: PROTEIN FIBRIL Ligands: LHG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-08-07 Classification: TRANSCRIPTION REGULATOR Ligands: MRD |
 |
Crystal Structure Of Pqsr Coinducer Binding Domain Of Pseudomonas Aeruginosa With Ligand Nhq
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-08-07 Classification: TRANSCRIPTION REGULATOR Ligands: NNQ |
 |
Crystal Structure Of Pqsr Co-Inducer Binding Domain Of Pseudomonas Aeruginosa With Inhibitor 3Nh2-7Cl-C9Qzn
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-08-07 Classification: TRANSCRIPTION REGULATOR/INHIBITOR Ligands: QZN |
 |
Structure Of A Translocation Signal Domain Mediating Conjugative Transfer By Type Iv Secretion Systems
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-06-19 Classification: HYDROLASE Ligands: SO4, MG |

