Search Count: 25
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2022-03-09 Classification: DNA |
 |
Organism: Streptomyces sp. nbrc 109436
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-09-01 Classification: BIOSYNTHETIC PROTEIN Ligands: CL, CAC, MG |
 |
Organism: Streptomyces sp. nbrc 109436
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-09-01 Classification: BIOSYNTHETIC PROTEIN Ligands: CAC, CL |
 |
Organism: Streptomyces sp. nbrc 109436
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-09-01 Classification: BIOSYNTHETIC PROTEIN/INHIBITOR Ligands: CL, ZHU |
 |
Organism: Listeria innocua serovar 6a (strain atcc baa-680 / clip 11262)
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2017-12-06 Classification: HYDROLASE/ANTIBIOTIC Ligands: DMS, 7YU |
 |
Crystal Structure Of Nad+-Reducing [Nife]-Hydrogenase In The Air-Oxidized State
Organism: Hydrogenophilus thermoluteolus
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2017-08-23 Classification: OXIDOREDUCTASE Ligands: FMN, SF4, FES, FCO, 3NI, MG |
 |
Crystal Structure Of Nad+-Reducing [Nife]-Hydrogenase In The H2-Reduced State
Organism: Hydrogenophilus thermoluteolus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-08-23 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FCO, NI, MG |
 |
The Structure Of Monooxygenase Ksta11 In The Biosynthetic Pathway Of Kosinostatin
Organism: Micromonospora sp. tp-a0468
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2016-12-21 Classification: OXIDOREDUCTASE Ligands: GOL, DTT |
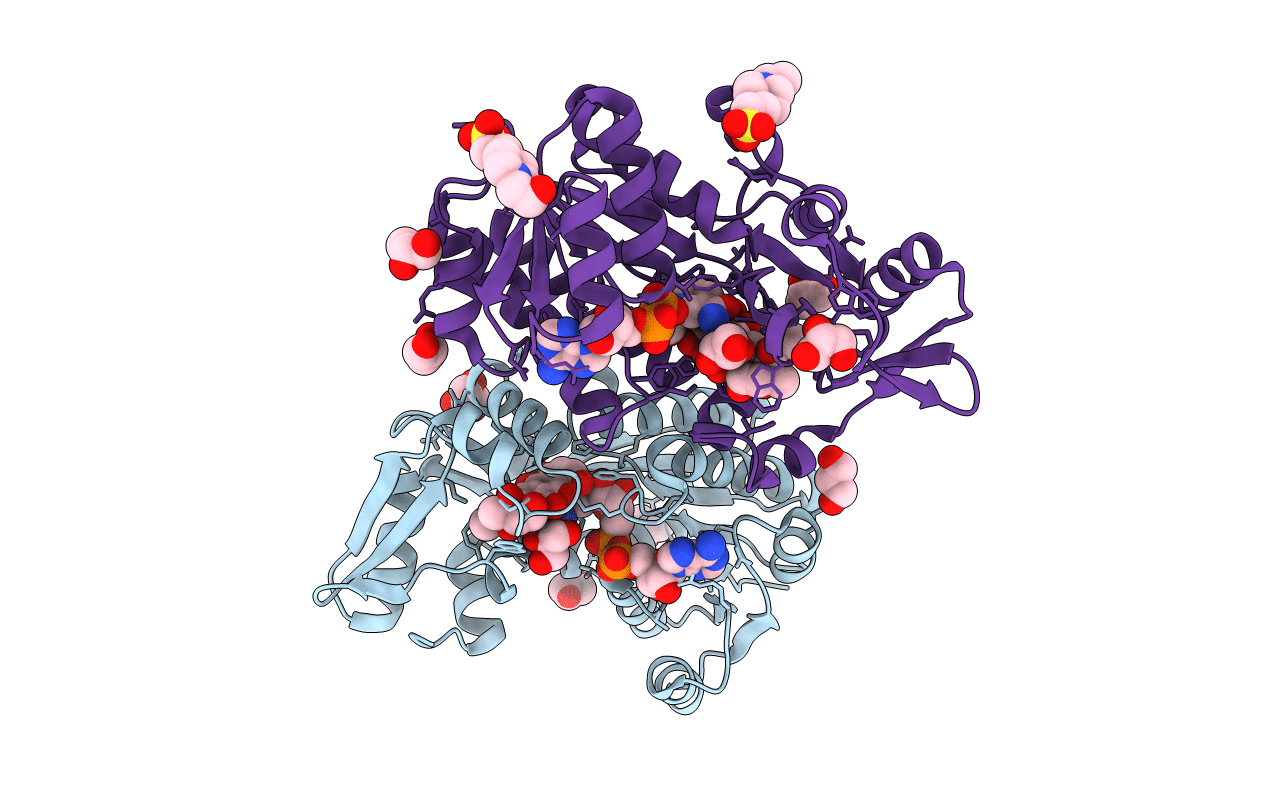 |
The Structure Of Monooxygenase Ksta11 In Complex With Nad And Its Substrate
Organism: Micromonospora sp. tp-a0468
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-12-21 Classification: OXIDOREDUCTASE Ligands: NAD, 5VD, GOL, EPE, MG |
 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing A 3-Methyladenine Analog (100% Substrate At 4 Hours)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA |
 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing A 3-Methyladenine Analog Or Dna Containing An Abasic Site And A Free Nucleobase (71% Substrate/29% Product At 24 Hours)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: 54K |
 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing A 3-Methyladenine Analog Or Dna Containing An Abasic Site And A Free Nucleobase (51% Substrate/49% Product At 48 Hours)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: 54K |
 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing A 3-Methyladenine Analog Or Dna Containing An Abasic Site And A Free Nucleobase (33% Substrate/67% Product At 72 Hours)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: 54K |
 |
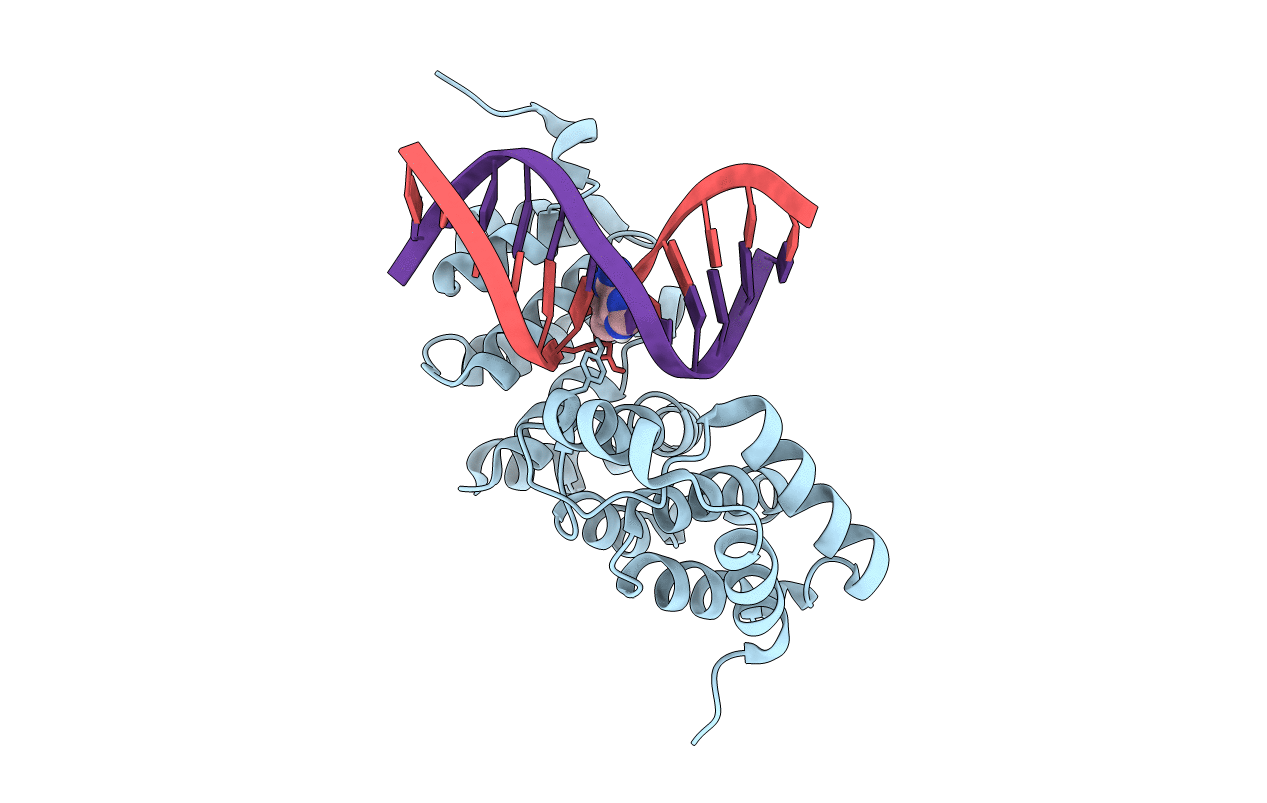
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing A 3-Methyladenine Analog Or Dna Containing An Abasic Site And A Free Nucleobase (18% Substrate/82% Product At 96 Hours)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: 54K |
 |
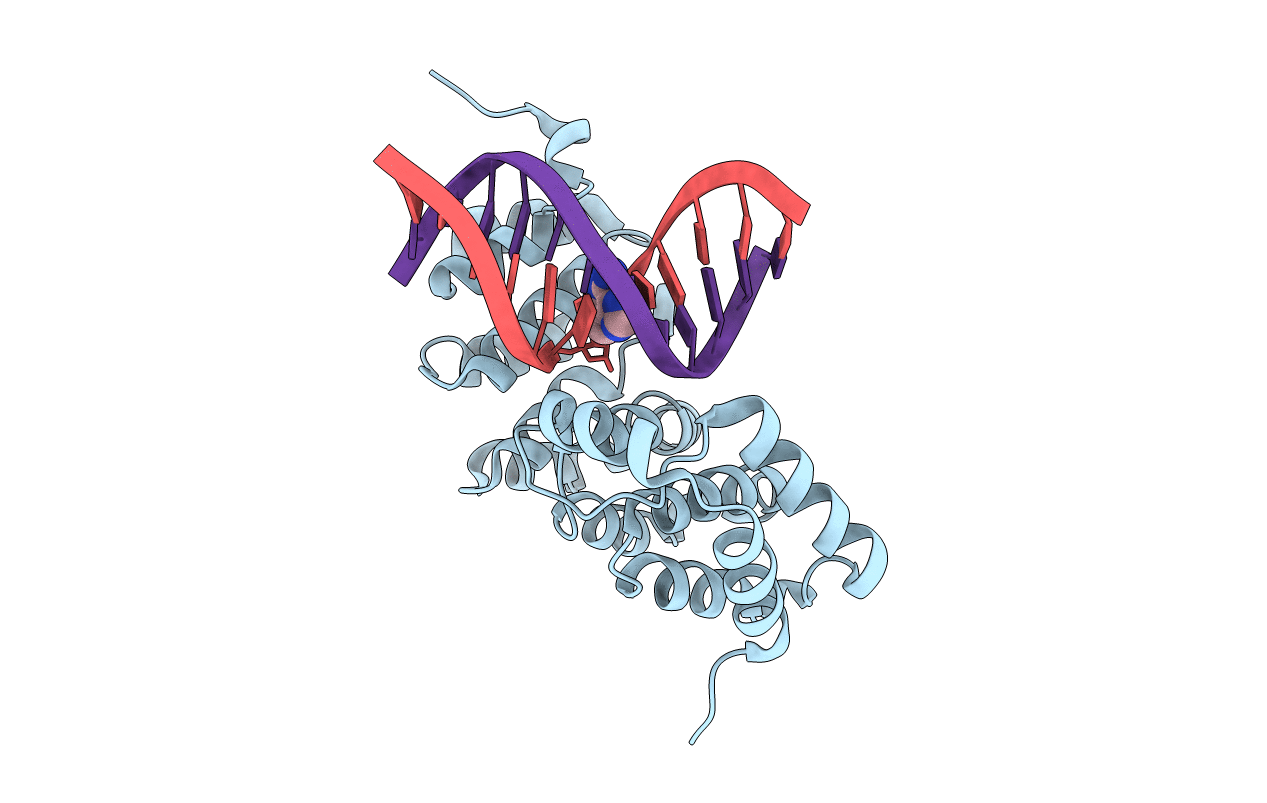
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing An Abasic Site And A Free Nucleobase (100% Product At 144 Hours)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: 54K |
 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing An Abasic Site And A Free Nucleobase (100% Product At 240 Hours)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: 54K |
 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing An Abasic Site And A Free Nucleobase (100% Product At 360 Hours)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: 54K |
 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing A 3-Methyladenine Analog (9-Mer A)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA |
 |
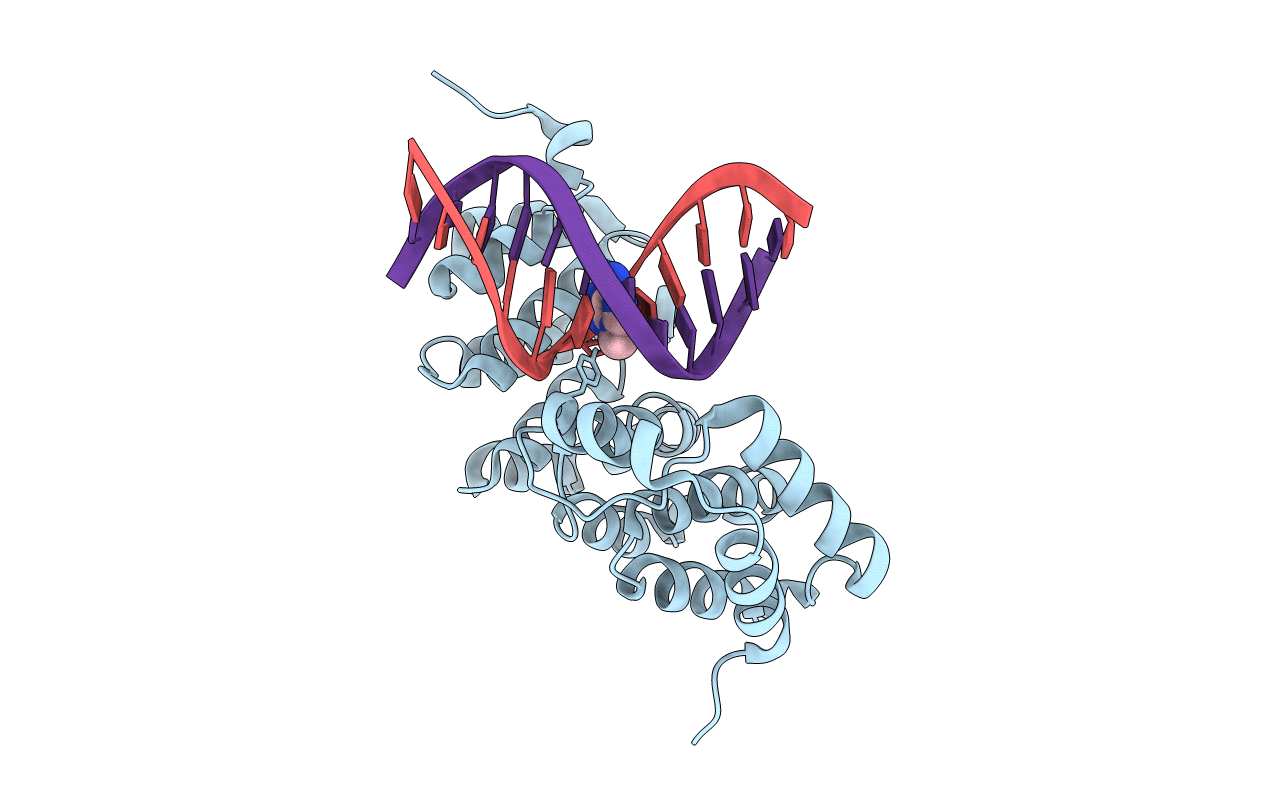
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing A 3-Methyladenine Analog (9-Mer B)
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA |
 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing An Oxocarbenium-Intermediate Analog And A Free 3-Methyladenine Nucleobase
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: ADK |

