Search Count: 27
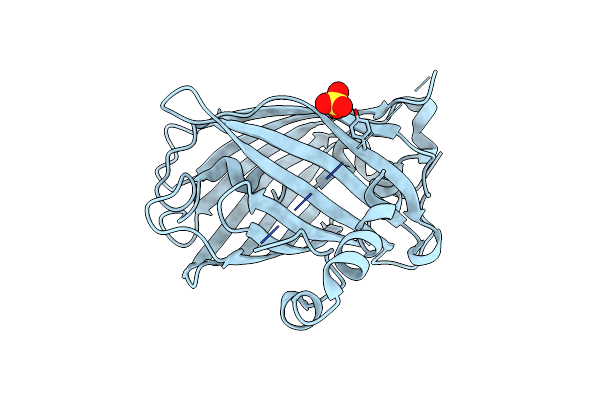 |
Crystal Structure Of Yeast Enhanced Green Fluorescent Protein - Mouse Polymerase Iota Ubiquitin Binding Motif Fusion Protein
Organism: Aequorea victoria, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-09-29 Classification: Fluorescent Protein, replication Ligands: SO4 |
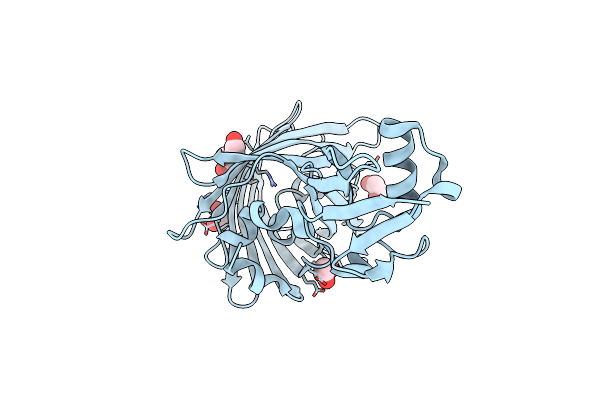 |
Crystal Structure Of Yeast Enhanced Green Fluorescent Protein-Ubiquitin Fusion Protein
Organism: Aequorea victoria, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-09-29 Classification: Fluorescent Protein, transcription Ligands: EDO |
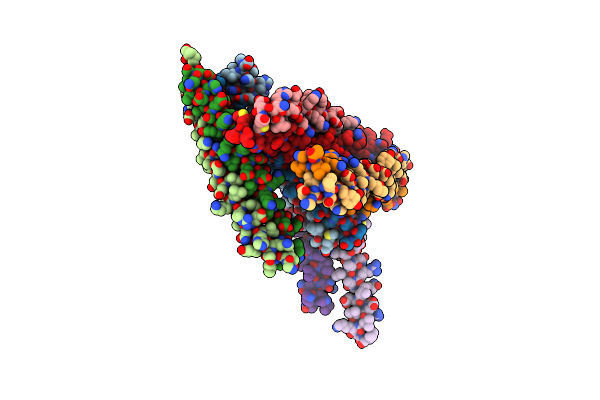 |
Crystal Structure Of The Head-To-Tail Junction Of Tropomyosin Complexed With A Fragment Of Tnt
Organism: Saccharomyces cerevisiae, Oryctolagus cuniculus, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2008-04-22 Classification: CONTRACTILE PROTEIN |
 |
Organism: Saccharomyces cerevisiae, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-22 Classification: CONTRACTILE PROTEIN Ligands: MG |
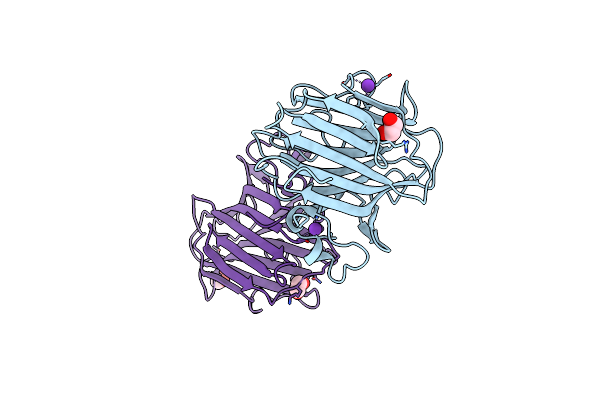 |
Crystal Structure Of Emp46P Carbohydrate Recognition Domain (Crd), Potassium-Bound Form
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2006-01-31 Classification: SUGAR BINDING PROTEIN Ligands: K, EDO |
 |
Crystal Structure Of Emp46P Carbohydrate Recognition Domain (Crd), Metal-Free Form
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2006-01-31 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Emp46P Carbohydrate Recognition Domain (Crd), Y131F Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2006-01-31 Classification: SUGAR BINDING PROTEIN Ligands: K, EDO |
 |
Crystal Structure Of Emp47P Carbohydrate Recognition Domain (Crd), Tetragonal Crystal Form
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2006-01-31 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Emp47P Carbohydrate Recognition Domain (Crd), Monoclinic Crystal Form 1
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2006-01-31 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Emp47P Carbohydrate Recognition Domain (Crd), Monoclinic Crystal Form 2
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2006-01-31 Classification: SUGAR BINDING PROTEIN Ligands: EDO |
 |
Crystal Structure Of Emp47P Carbohydrate Recognition Domain (Crd), Orthorhombic Crystal Form
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-01-31 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of The T. Thermophilus Rna Polymerase Holoenzyme In Complex With Inhibitor Tagetitoxin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-11-08 Classification: TRANSFERASE Ligands: MG, ZN, TGT |
 |
Crystal Structure Of The T. Thermophilus Rna Polymerase Holoenzyme In Complex With Antibiotic Rifabutin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-09-20 Classification: TRANSFERASE Ligands: MG, RBT, ZN |
 |
Crystal Structure Of The T. Thermophilus Rna Polymerase Holoenzyme In Complex With Antibiotic Rifapentin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-09-20 Classification: TRANSFERASE Ligands: MG, RPT, ZN |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-09-20 Classification: TRANSFERASE Ligands: ZN, MG |
 |
Crystal Structure Of The T. Thermophilus Rna Polymerase Holoenzyme In Complex With Antibiotic Sterptolydigin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-09-20 Classification: TRANSFERASE Ligands: STD, ZN, MG |
 |
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-06-28 Classification: PROTEIN TRANSPORT/SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-05-20 Classification: PROTEIN TRANSPORT |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2003-05-20 Classification: PROTEIN TRANSPORT Ligands: MG, GTP |
 |
Crystal Structure Of Gga1 Gat N-Terminal Region In Complex With Arf1 Gtp Form
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2003-05-06 Classification: PROTEIN TRANSPORT Ligands: MG, IOD, GTP |

