Search Count: 21
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz8 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, MG |
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz8 In Complex With Pap, Involved In Caprazamycin Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, A3P, SO4 |
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz4 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, MG |
 |
Crystal Structure Of The Reaction Intermediate Of Streptomyces Sp. Sulfotransferase Cpz4 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, MG |
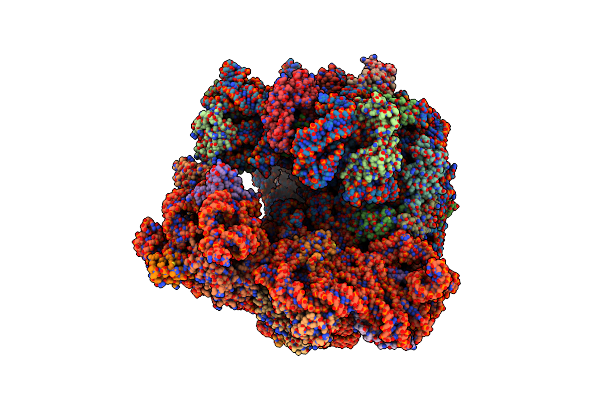 |
Organism: Mycobacterium tuberculosis variant bovis bcg str. pasteur 1173p2
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME/ANTIBIOTIC Ligands: ZN, MG, A1L32 |
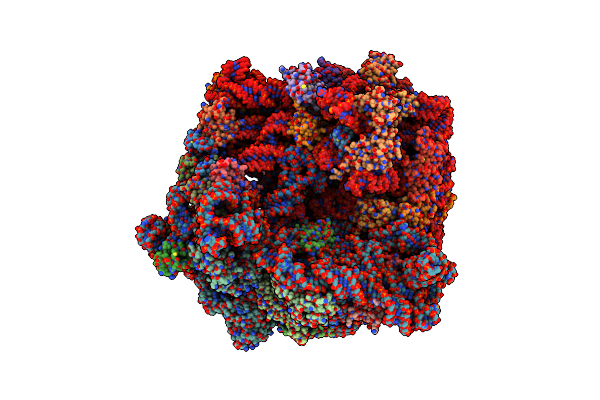 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: RIBOSOME Ligands: MG, SPD, 84D, SPM, ZN |
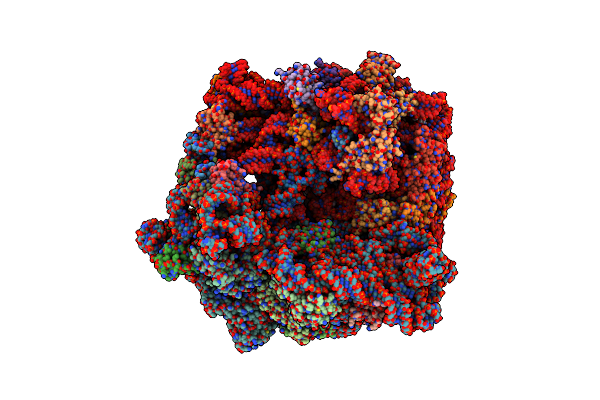 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: RIBOSOME Ligands: MG, SPD, 84G, SPM, ZN |
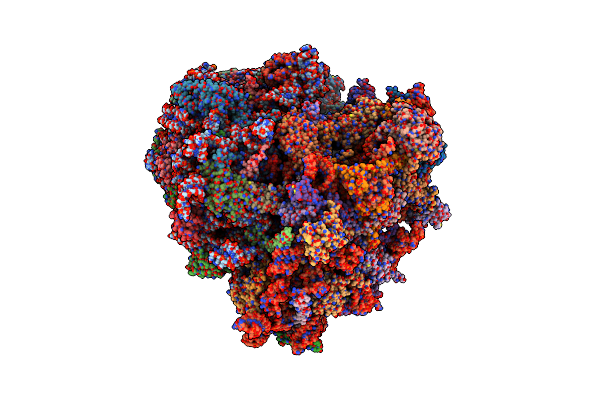 |
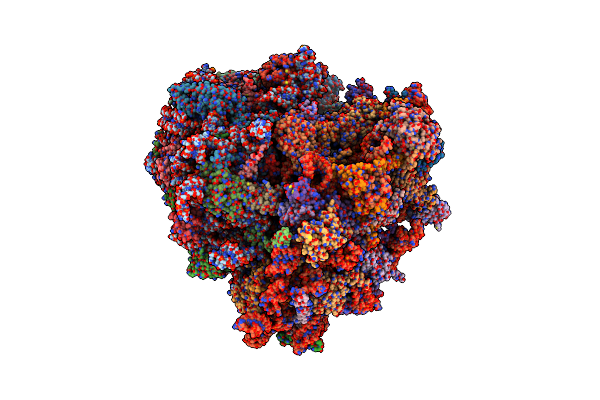
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: RIBOSOME Ligands: 84D, MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: RIBOSOME Ligands: 84G, MG, ZN |
 |
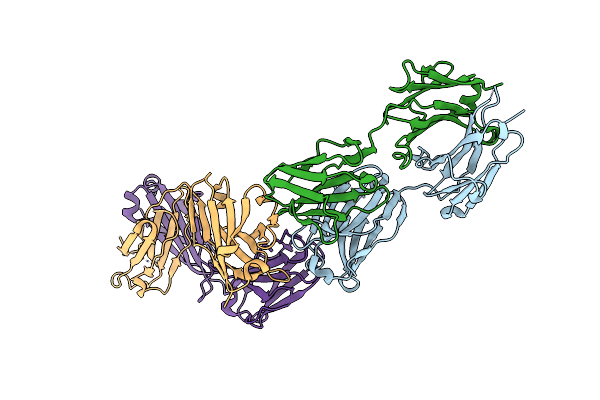
Organism: Lake victoria marburgvirus (strain angola/2005), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-03-09 Classification: VIRAL PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-09-26 Classification: IMMUNE SYSTEM |
 |
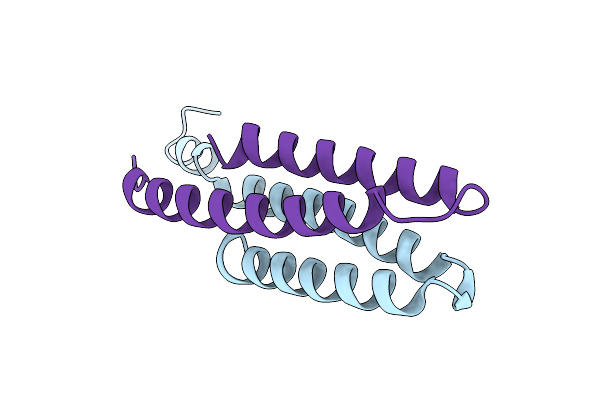
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2016-12-14 Classification: SIGNALING PROTEIN |
 |
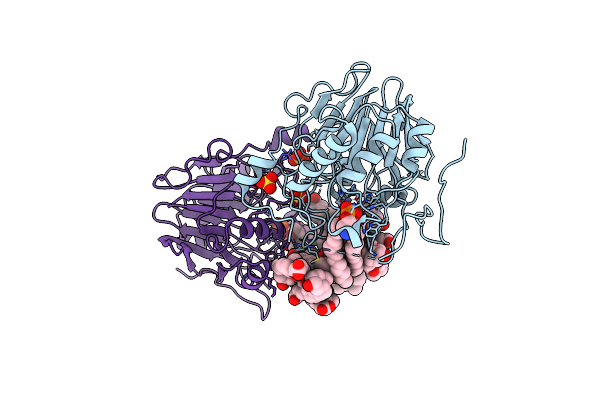
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-12-14 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-06-17 Classification: HYDROLASE Ligands: ZN, 3PE, DXC, SO4 |
 |
Organism: Toxoplasma gondii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-10-15 Classification: LYASE Ligands: BME, SO4, ACT, GOL |
 |
1.37 Angstrom Resolution Crystal Structure Of Apo-Form Of A Putative Deoxyribose-Phosphate Aldolase From Toxoplasma Gondii Me49
Organism: Toxoplasma gondii
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2012-05-23 Classification: LYASE Ligands: CL, BME |
 |
1.8 Angstrom Resolution Crystal Structure Of A Putative Deoxyribose-Phosphate Aldolase From Toxoplasma Gondii Me49
Organism: Toxoplasma gondii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-03-30 Classification: LYASE Ligands: SO4 |
 |
Crystal Structure Of Rat Vitamin D Receptor Bound To Adamantyl Vitamin D Analogs: Structural Basis For Vitamin D Receptor Antagonism And/Or Partial Agonism
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-09-02 Classification: TRANSCRIPTION Ligands: NYA |
 |
Crystal Structure Of Rat Vitamin D Receptor Bound To Adamantyl Vitamin D Analogs: Structural Basis For Vitamin D Receptor Antagonism And/Or Partial Agonism
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-09-02 Classification: TRANSCRIPTION Ligands: TT2, EDO, FMT |
 |
Crystal Structure Of Rat Vitamin D Receptor Bound To Adamantyl Vitamin D Analogs: Structural Basis For Vitamin D Receptor Antagonism And/Or Partial Agonism
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2008-09-02 Classification: TRANSCRIPTION Ligands: MI4 |

