Search Count: 44
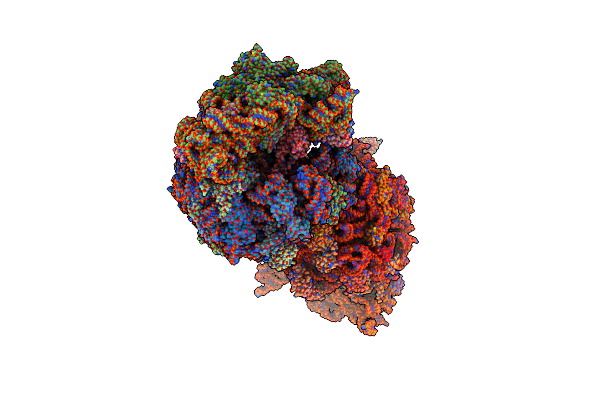 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Fva1 Antimicrobial Peptide, Mrna, A-Site Release Factor 1, And Deacylated P-Site And E-Site Trnaphe At 2.70A Resolution
Organism: Thermus thermophilus hb8, Escherichia coli, Escherichia phage t4, Frieseomelitta varia
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-11-13 Classification: RIBOSOME Ligands: MG, ZN, SF4 |
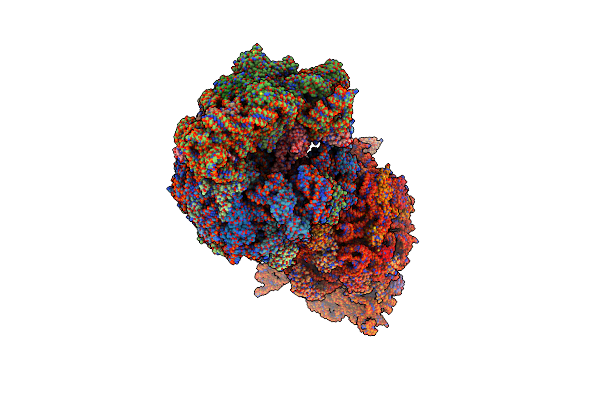 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Api Antimicrobial Peptide, Mrna, A-Site Release Factor 1, And Deacylated P-Site And E-Site Trnaphe At 2.85A Resolution
Organism: Thermus thermophilus hb8, Escherichia coli, Escherichia phage t4, Apis mellifera
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-11-13 Classification: RIBOSOME Ligands: MG, ZN, SF4 |
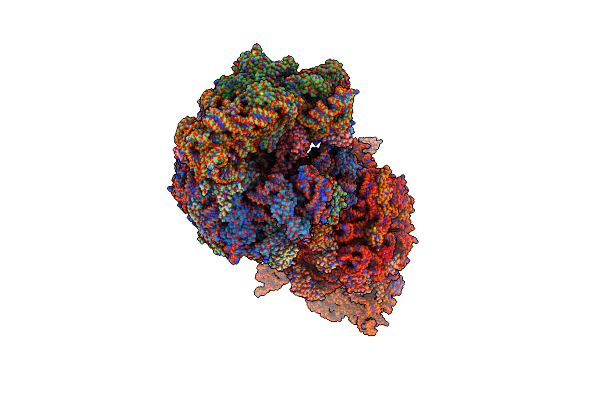 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Api137 Antimicrobial Peptide, Mrna, A-Site Release Factor 1, And Deacylated P-Site And E-Site Trnaphe At 2.70A Resolution
Organism: Thermus thermophilus hb8, Escherichia coli, Escherichia phage t4, Apis mellifera
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-11-13 Classification: RIBOSOME Ligands: MG, ZN, SF4 |
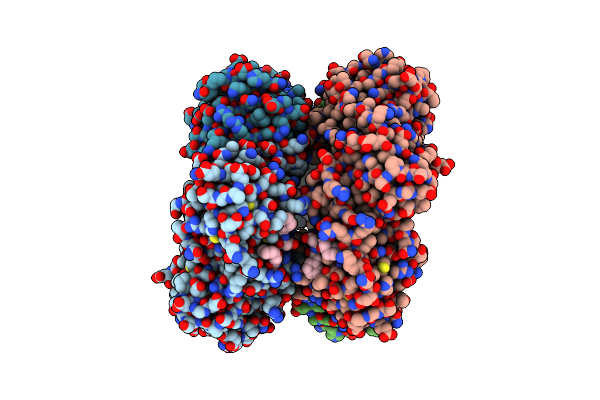 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: PG4, PEG |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: PEG, GOL, ACT, CL |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: FMT, 1PE |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: PEG, 1PE, GOL |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: GOL |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: 1PE, PEG |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: MLI, GOL, 1PE |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: GOL, PEG |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: GOL, K, 1PE, CL, PEG |
 |
Organism: Escherichia coli 2-427-07_s4_c3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: GOL, PEG |
 |
Organism: Alistipes
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-02 Classification: HYDROLASE |
 |
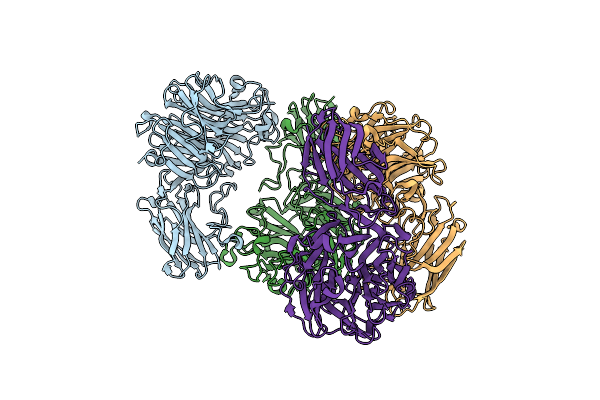
Organism: Unidentified bacterium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: DAN |
 |
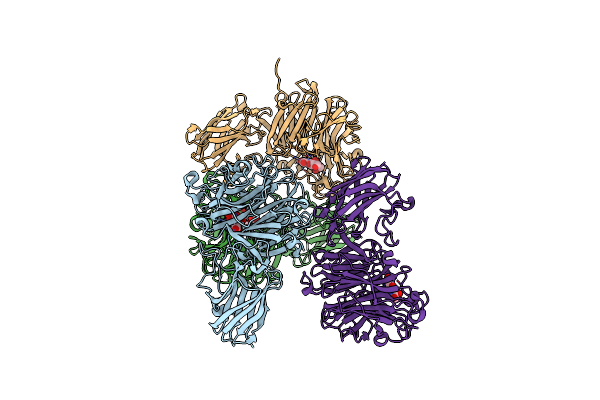
Organism: Unidentified bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-02 Classification: HYDROLASE |
 |
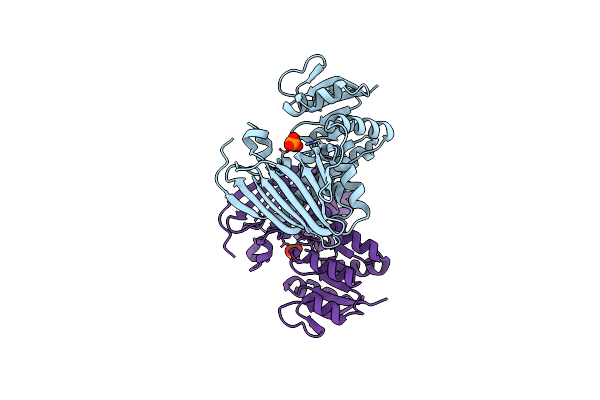
Organism: Unidentified bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: GC9 |
 |
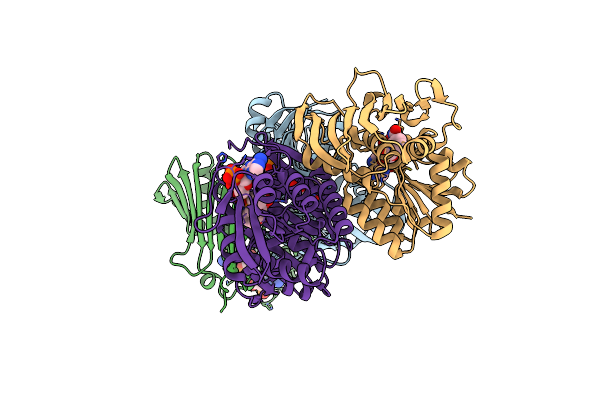
Crystal Structure Of Azospirillum Brasilense L-Arabinose 1-Dehydrogenase (Apo-Form)
Organism: Azospirillum brasilense
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Crystal Structure Of Azospirillum Brasilense L-Arabinose 1-Dehydrogenase (Nadp-Bound Form)
Organism: Azospirillum brasilense
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: NAP |

