Search Count: 18
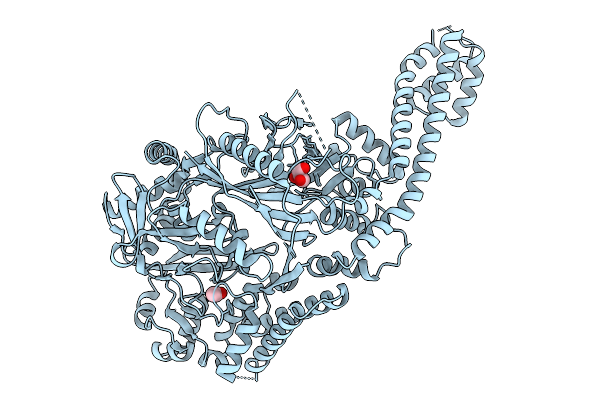 |
Crystal Structure Of Godf, A Post-Translational Modification Enzyme Involved In The Biosynthesis Of Goadsporin
Organism: Streptomyces sp. tp-a0584
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE Ligands: ACT, GOL |
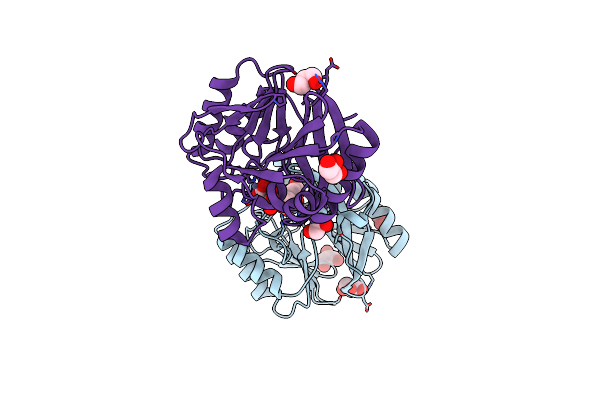 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: FLC, ZN, GOL |
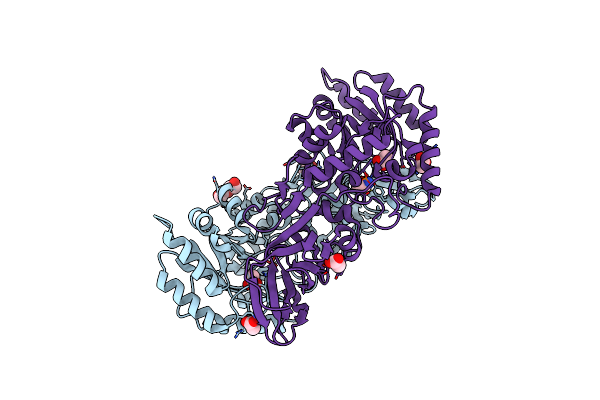 |
Organism: Latilactobacillus sakei
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-03-01 Classification: ISOMERASE Ligands: ACT, PEG, ALA |
 |
Crystal Structure Of Metallo-Beta-Lactamase Imp-27 From Morganella Morganii
Organism: Morganella morganii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Mox-1 Complexed With A Boronic Acid Transition State Inhibitor S02030
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: ZXM, ACT, ZN |
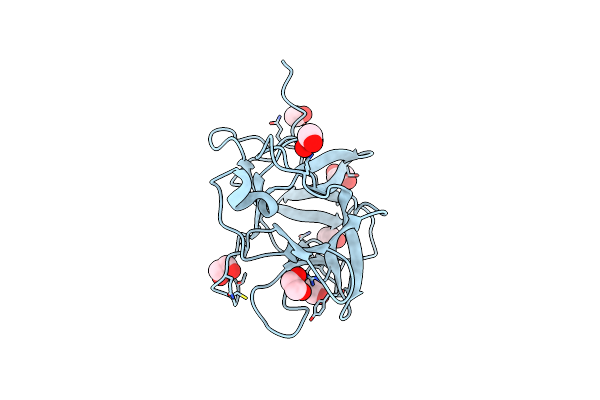 |
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-10-03 Classification: HYDROLASE INHIBITOR Ligands: EDO |
 |
Crystal Structure Of Metallo-Beta-Lactamase Imp-18 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-15 Classification: HYDROLASE Ligands: ZN, CIT |
 |
Crystal Structure Of Class C Beta-Lactamase Mox-1 Covalently Complexed With Aztorenam
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-07-01 Classification: HYDROLASE Ligands: AZR, ZN, ACT |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: ZN, ACT |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-23 Classification: HYDROLASE |
 |
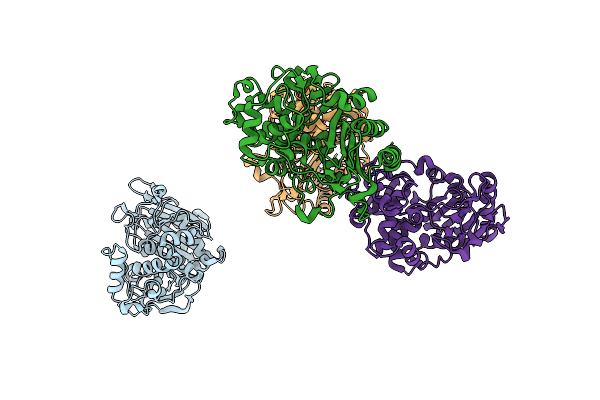
Crystal Structure Analysis Of Pyruvate Kinase From Bacillus Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-02-05 Classification: TRANSFERASE Ligands: SO4 |
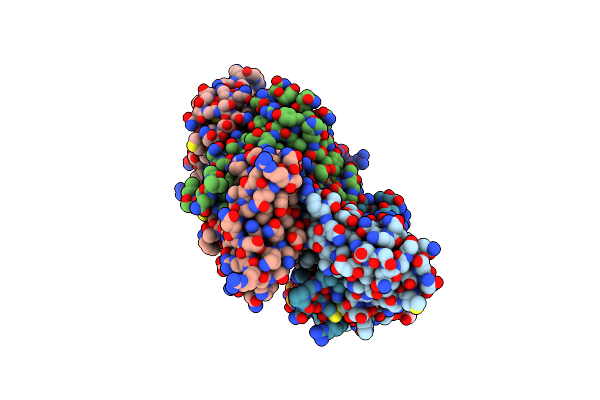 |
Crystal Structure Of Neoculin, A Sweet Protein With Taste-Modifying Activity.
Organism: Curculigo latifolia
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2006-06-20 Classification: PLANT PROTEIN Ligands: NAG |
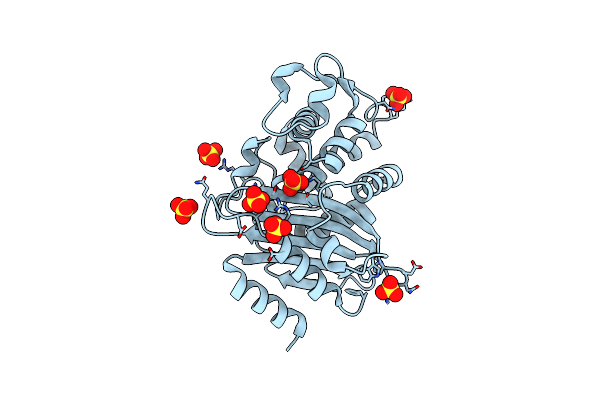 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2005-03-15 Classification: HYDROLASE Ligands: SO4 |
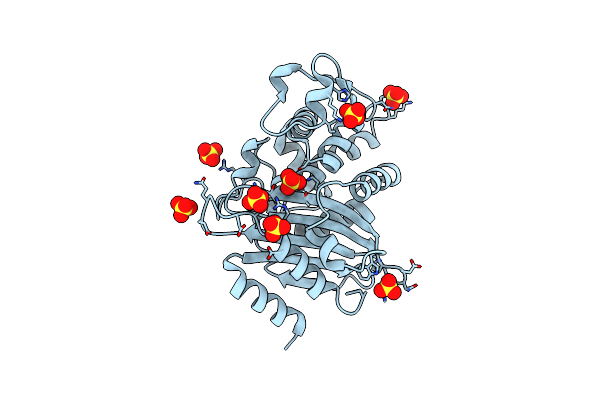 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2003-10-14 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-12-11 Classification: HYDROLASE Ligands: SO4, CEF |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-12-11 Classification: HYDROLASE Ligands: SO4, CEP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-12-11 Classification: HYDROLASE Ligands: SO4, PNM |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1999-04-27 Classification: HYDROLASE Ligands: SO4 |

