Search Count: 16
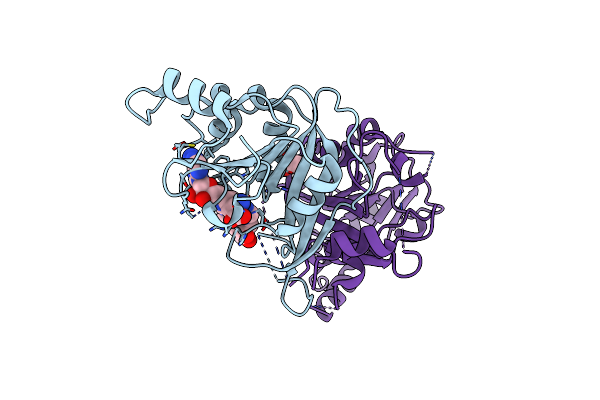 |
Crystal Structure Of The Human Mettl3-Mettl14 In Complex With A Bisubstrate Analogue (Ba2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: HZ2, ACT |
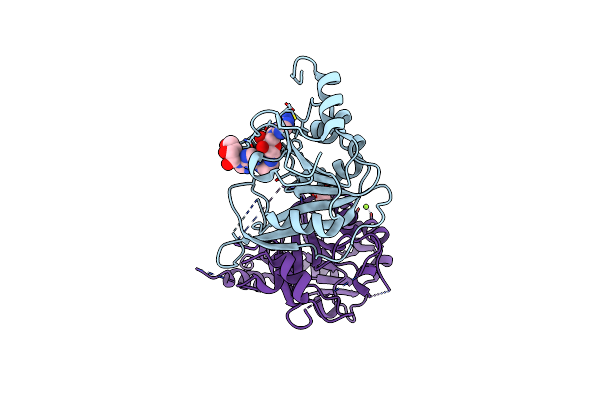 |
Crystal Structure Of The Human Mettl3-Mettl14 In Complex With A Bisubstrate Analogue (Ba1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: H9D, ACT, MG |
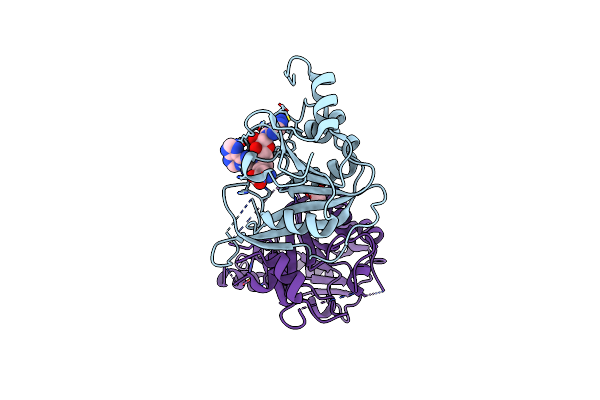 |
Crystal Structure Of The Human Mettl3-Mettl14 In Complex With A Bisubstrate Analogue (Ba4)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: I0C, ACT, MG |
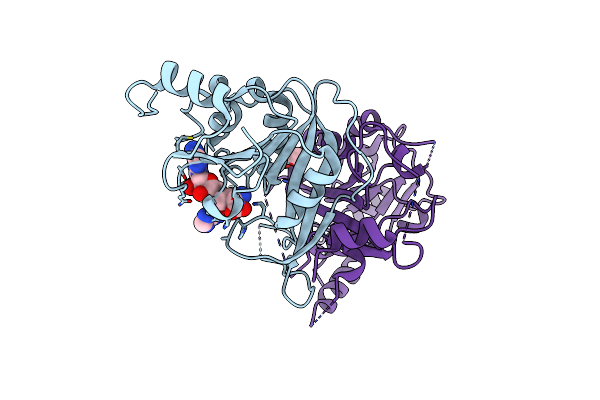 |
Crystal Structure Of The Human Mettl3-Mettl14 In Complex With A Bisubstrate Analogue (Ba6)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: H3I, ACT |
 |
Crystal Structure Of Weissella Viridescens Femxvv Non-Ribosomal Amino Acid Transferase In Complex With A Peptidyl-Xna Conjugate
Organism: Weissella viridescens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-10-26 Classification: TRANSFERASE Ligands: GOL, UDP, MUB |
 |
Crystal Structure Of Weissella Viridescens Femxvv Non-Ribosomal Amino Acid Transferase In Complex With A Peptidyl-Xna Conjugate
Organism: Weissella viridescens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2022-10-26 Classification: TRANSFERASE Ligands: GOL, UDP, MUB |
 |
Crystal Structure Of Weissella Viridescens Femxvv Non-Ribosomal Amino Acid Transferase In Complex With A Peptidyl-Xna Conjugate
Organism: Weissella viridescens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-10-26 Classification: TRANSFERASE Ligands: GOL, UDP, MUB |
 |
Crystal Structure Of Weissella Viridescens Femxvv Non-Ribosomal Amino Acid Transferase In Complex With A Peptidyl-Xna Conjugate
Organism: Weissella viridescens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-10-26 Classification: TRANSFERASE Ligands: GOL, UDP, MUB |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-06-01 Classification: RNA BINDING PROTEIN Ligands: 6D6 |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-06-01 Classification: RNA BINDING PROTEIN Ligands: 6D6 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-05-25 Classification: RNA BINDING PROTEIN Ligands: 0Y0 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: HY8 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: HZ2 |
 |
Structure Of E.Coli Rlmj In Complex With The Natural Cofactor Product S-Adenosyl-Homocysteine (Sah)
Organism: Escherichia coli pcn033
Method: X-RAY DIFFRACTION Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: SAH |
 |
Structure Of M. Capricolum Trmk In Complex With The Natural Cofactor Product S-Adenosyl-Homocysteine (Sah)
Organism: Mycoplasma capricolum subsp. capricolum
Method: X-RAY DIFFRACTION Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: P3G, NXL, GOL, CL |

