Search Count: 77
 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-05-02 Classification: ISOMERASE,Oxidoreductase Ligands: MRD, MN, CL, CA |
 |
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 1997-06-16 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PO4 |
 |
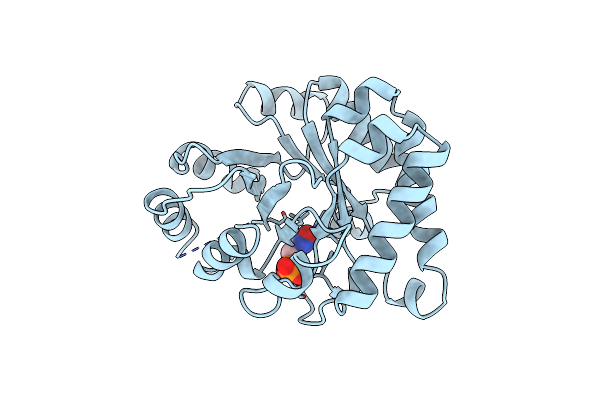
Three New Crystal Structures Of Point Mutation Variants Of Monotim: Conformational Flexibility Of Loop-1,Loop-4 And Loop-8
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1995-10-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGA |
 |
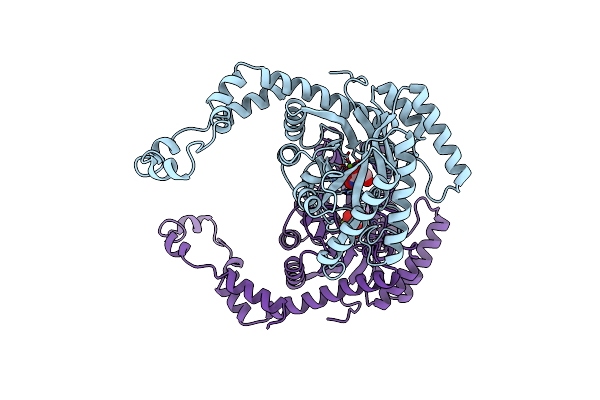
Three New Crystal Structures Of Point Mutation Variants Of Monotim: Conformational Flexibility Of Loop-1,Loop-4 And Loop-8
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1995-09-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGH |
 |
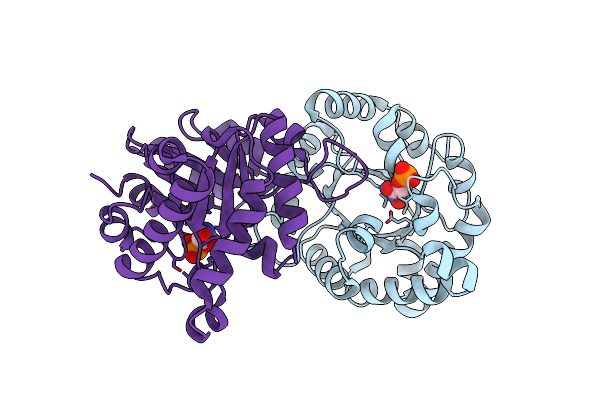
Design, Synthesis, And Characterization Of A Potent Xylose Isomerase Inhibitor, D-Threonohydroxamic Acid, And High-Resolution X-Ray Crystallographic Structure Of The Enzyme-Inhibitor Complex
Organism: Streptomyces olivochromogenes
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1995-07-10 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: MG, HYA |
 |
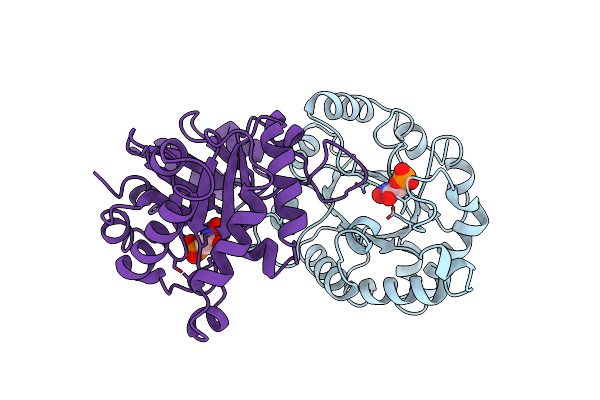
S96P Change Is A Second-Site Suppressor For H95N Sluggish Mutant Triosephosphate Isomerase
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-04-20 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGH |
 |
S96P Change Is A Second-Site Suppressor For H95N Sluggish Mutant Triosephosphate Isomerase
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-04-20 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGH |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-04-20 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGH |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-02-14 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGH |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-02-14 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGH |
 |
Crystal Structure Of Recombinant Human Triosephosphate Isomerase At 2.8 Angstroms Resolution. Triosephosphate Isomerase Related Human Genetic Disorders And Comparison With The Trypanosomal Enzyme
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1995-01-26 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGA |
 |
Large Scale Structural Rearrangements Of The Front Loops In Monomerised Triosephosphate Isomerase, As Deduced From The Comparison Of The Structural Properties Of Monotim And Its Point Mutation Variant Monoss
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-09-30 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) |
 |
The Crystal Structure Of An Engineered Monomeric Triosephosphate Isomerase, Monotim: The Correct Modelling Of An Eight-Residue Loop
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-07-31 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: SO4 |
 |
Modular Mutagenesis Of A Tim-Barrel Enzyme: The Crystal Structure Of A Chimeric E. Coli Tim Having The Eighth (Beta-Alpha)-Unit Replaced By The Equivalent Unit Of Chicken Tim
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: SO4 |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: MN |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XLS, MN |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: MN, ASC |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: ASO, MN |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: GLC, MN |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XYL, MN |

