Search Count: 13
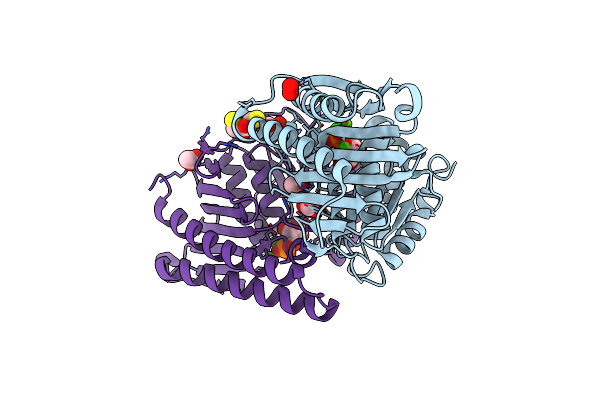 |
Crystal Structure Of Pseudomonas Aeruginosa Suhb In Complex With D-Myo-Inositol-1-Phosphate
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: IPD, ACT, PEO, GOL, DTU, CL, CA |
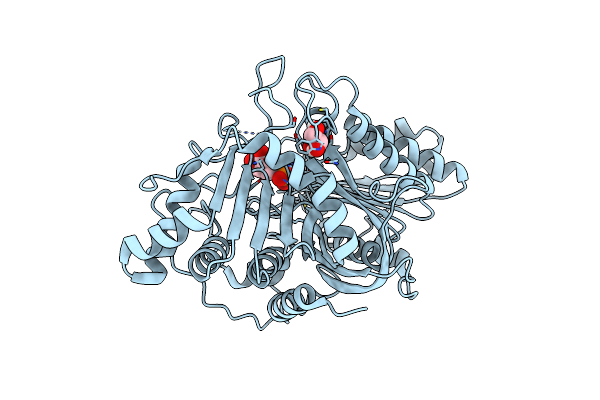 |
Phospholipase D Engineered Mutant (Tnyr) Inactive Enzyme (H168A) Bound To 1-Inositol Phosphate
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-28 Classification: LIPID BINDING PROTEIN Ligands: IPD, MES, GOL |
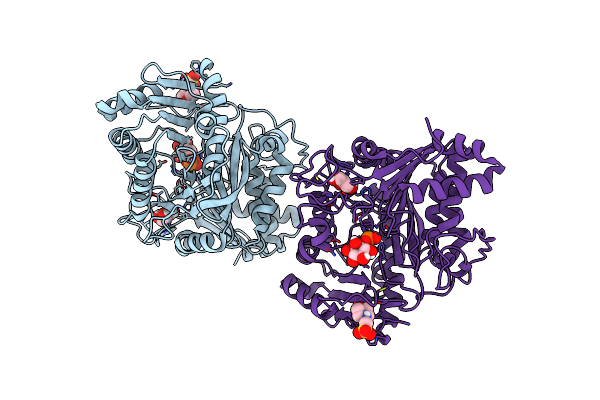 |
Phospholipase D Engineered Mutant (Tnyr) H442 Covalent Adduct With 1-Inositol Phosphate
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: GOL, MES, IPD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-03-25 Classification: SUGAR BINDING PROTEIN Ligands: CA, B3P, MAN, IPD |
 |
Organism: Staphylococcus aureus (strain mssa476)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-02-08 Classification: HYDROLASE Ligands: CA, IPD, PO4 |
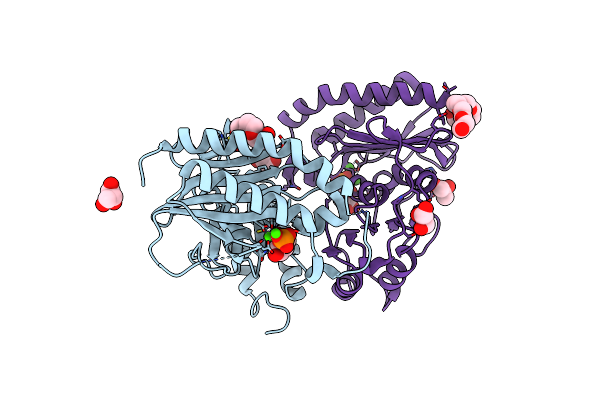 |
Crystal Structure Of Dual Specific Impase/Nadp Phosphatase Bound With D-Inositol-1-Phosphate
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: CA, PG4, CL, GOL, IPD, P6G |
 |
Structural Insights Into Substrate Binding Of Brown Spider Venom Class Ii Phospholipases D
Organism: Loxosceles intermedia
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-06-03 Classification: HYDROLASE Ligands: MG, PEG, IPD, GOL, OCA, SHV, DKA, PLM, TDA |
 |
Crystal Structure Of Inositol Phosphate Bound Trimeric Human Lung Surfactant Protein D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-11-17 Classification: SUGAR BINDING PROTEIN Ligands: CA, IPD |
 |
Crystal Structure Of The Trimeric Neck And Carbohydrate Recognition Domain Of Human Surfactant Protein D In Complex With Inositol-1-Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2007-05-08 Classification: SUGAR BINDING PROTEIN Ligands: CA, IPD |
 |
Crystal Structure Of A Ternary Complex Of Dual Activity Fbpase/Impase (Af2372) From Archaeoglobus Fulgidus With Calcium Ions And D-Myo-Inositol-1-Phosphate
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2002-05-22 Classification: HYDROLASE Ligands: CA, IPD |
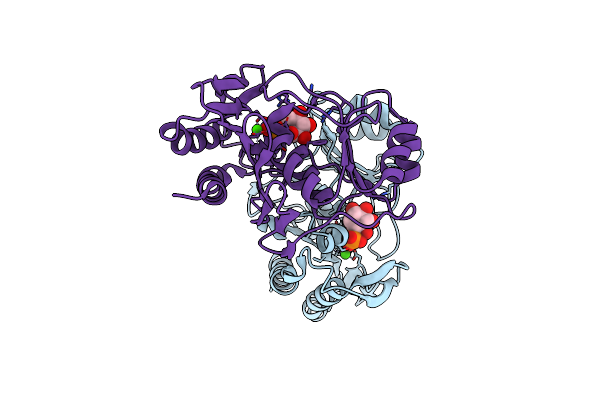 |
Crystal Structure Of Mj0109 Gene Product Inositol Monophosphatase-Fructose 1,6 Bisphosphatase
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2001-03-14 Classification: HYDROLASE Ligands: CA, IPD |
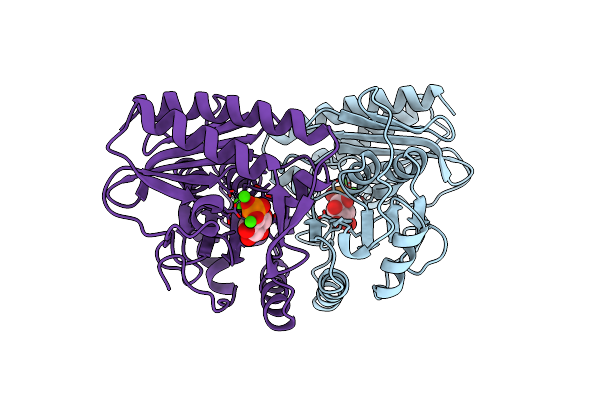 |
Human Myo-Inositol Monophosphatase In Complex With D-Inositol-1-Phosphate And Calcium
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1998-01-14 Classification: HYDROLASE Ligands: CA, CL, IPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1995-02-27 Classification: HYDROLASE Ligands: GD, IPD |

