Search Count: 57
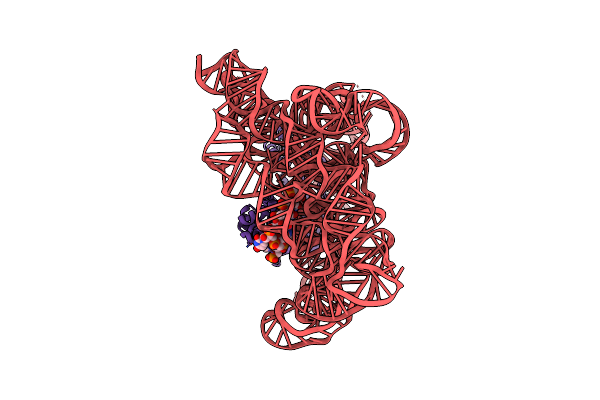 |
Structure Of A Group Ii Intron Ribonucleoprotein In The Pre-Ligation (Pre-2F) State
Organism: [eubacterium] rectale
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: Transferase/RNA Ligands: CA, NH4 |
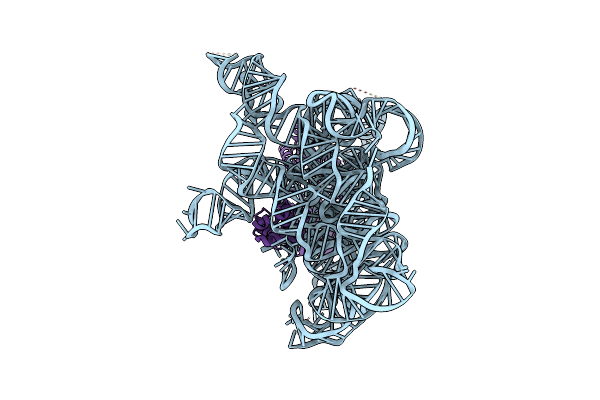 |
Structure Of A Group Ii Intron Ribonucleoprotein In The Pre-Branching (Pre-1F) State
Organism: [eubacterium] rectale
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: Transferase/RNA Ligands: CA, NH4 |
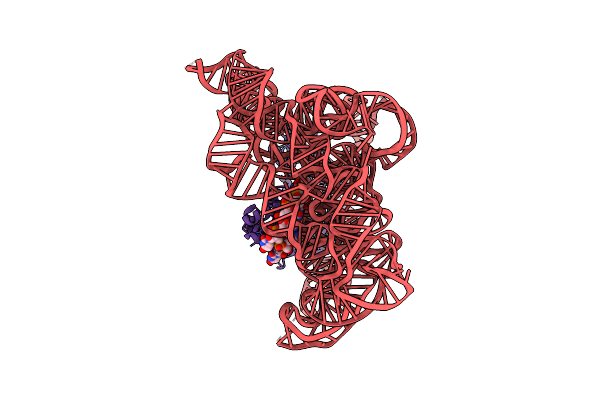 |
Structure Of A Group Ii Intron Ribonucleoprotein In The Post-Ligation (Post-2F) State
Organism: [eubacterium] rectale
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: Transferase/RNA Ligands: MG, NH4 |
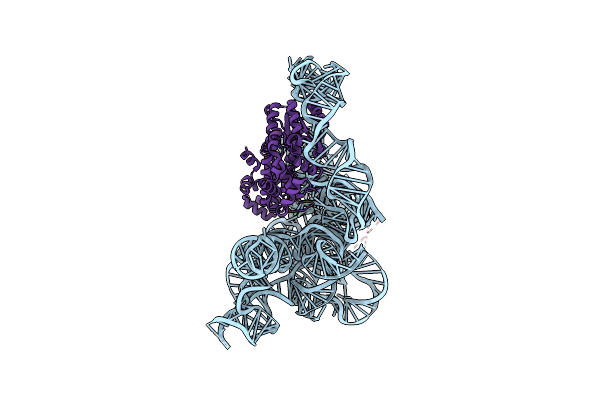 |
Organism: [eubacterium] rectale
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: RNA BINDING PROTEIN/RNA Ligands: MG, NH4 |
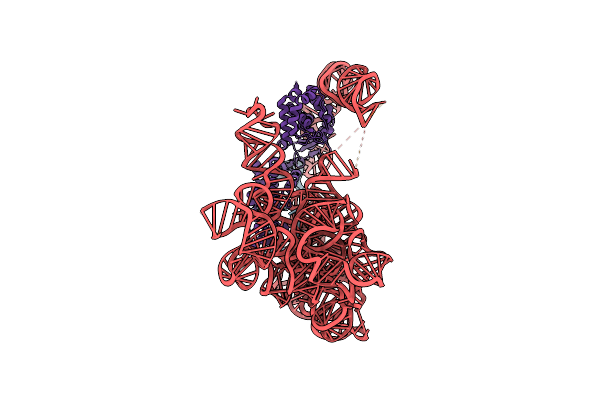 |
Organism: [eubacterium] rectale
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, NH4 |
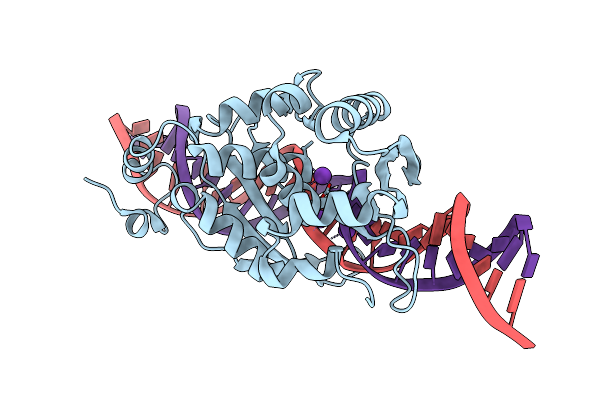 |
Organism: Wickerhamomyces canadensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: K |
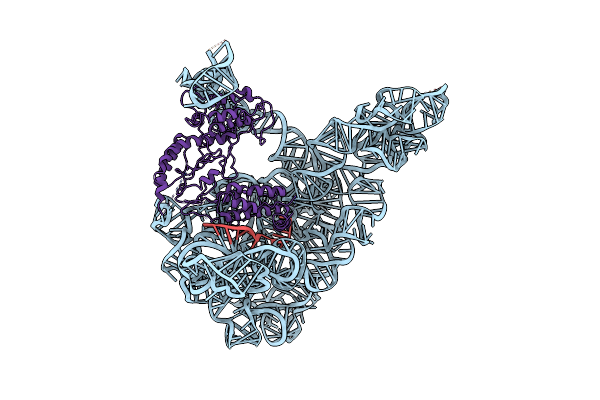 |
Cryo-Em Structure Of A Group Ii Intron Rnp Complexed With Its Reverse Transcriptase
Organism: Lactococcus lactis subsp. cremoris
Method: ELECTRON MICROSCOPY Release Date: 2020-10-21 Classification: TRANSFERASE/RNA |
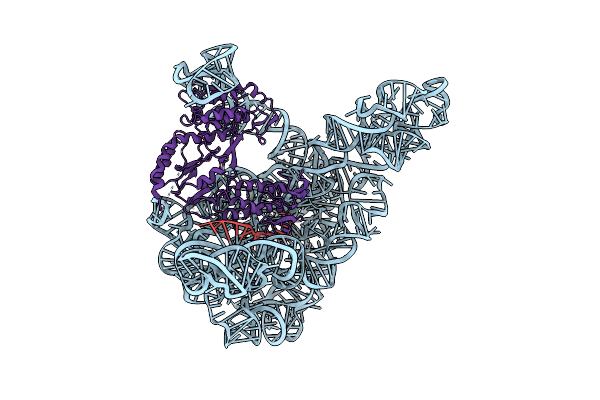 |
Organism: Lactococcus lactis, Lactococcus lactis subsp. cremoris
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2016-05-11 Classification: TRANSFERASE |
 |
The Crystal Structure Of I-Scei In Complex With Its Target Dna In The Presence Of Mn
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-05-04 Classification: HYDROLASE/DNA Ligands: MN |
 |
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Resolution:4.50 Å Release Date: 2016-05-04 Classification: TRANSFERASE |
 |
Expanding Lagalidadg Endonuclease Scaffold Diversity By Rapidly Surveying Evolutionary Sequence Space
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-02-29 Classification: HYDROLASE/DNA Ligands: CA, PEG, SO4, NA |
 |
Crystal Structure Of Y2 I-Anii Variant (F13Y/S111Y)/Dna Complex With Calcium
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-01-13 Classification: HYDROLASE/DNA Ligands: CA |
 |
Organism: Vulcanisaeta distributa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-12-30 Classification: HYDROLASE/DNA Ligands: MG, EOH |
 |
Organism: Thermoproteus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2006-07-06 Classification: HYDROLASE Ligands: CL, SO4 |
 |
Organism: Chlamydomonas eugametos, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-05-23 Classification: Hydrolase/DNA Ligands: CA |
 |
L116A Mutant Of The Homing Endonuclease I-Ppoi Complexed To Homing Site Dna.
Organism: Physarum polycephalum
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2000-08-03 Classification: Hydrolase/DNA Ligands: MG, ZN |
 |
Organism: Physarum polycephalum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-08-02 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Desulfurococcus mobilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-03-24 Classification: INTRON-ENCODED |
 |
Organism: Physarum polycephalum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1998-10-14 Classification: HYDROLASE/DNA Ligands: MG, ZN |
 |
Organism: Physarum polycephalum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1998-06-22 Classification: HYDROLASE/DNA Ligands: ZN |

