Search Count: 38
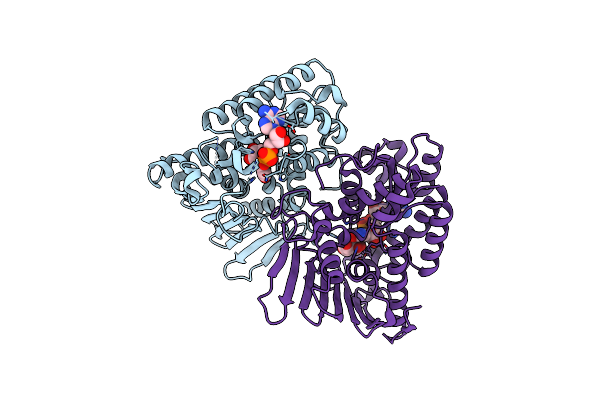 |
Crystal Structure Of Inositol Dehydrogenase Homolog Complexed With Nadh And Myo-Inositol From Azotobacter Vinelandii
Organism: Azotobacter vinelandii (strain dj / atcc baa-1303)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: NAI, INS |
 |
Inositol-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, INS |
 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase R178A Mutant, Complexed With Nad And Myo-Inositol, From Paracoccus Laeviglucosivorans
Organism: Paracoccus laeviglucosivorans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: ACT, NAD, INS |
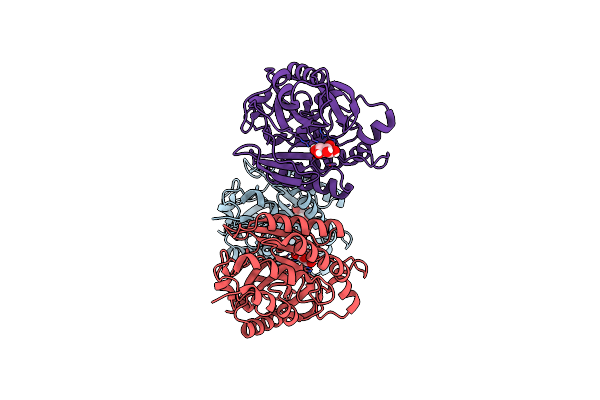 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-07-31 Classification: LIPID BINDING PROTEIN Ligands: INS |
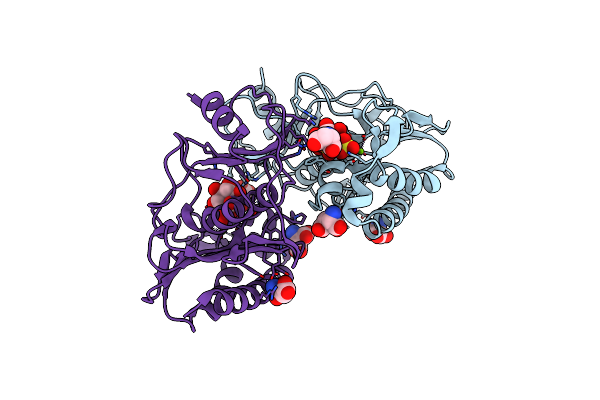 |
Organism: Archaeoglobus fulgidus (strain atcc 49558 / vc-16 / dsm 4304 / jcm 9628 / nbrc 100126)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-10-03 Classification: HYDROLASE Ligands: SO4, MG, ASP, INS |
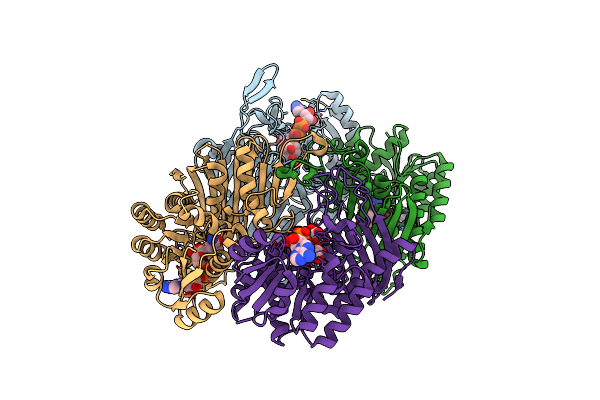 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase With L-Glucose Dehydrogenase Activity Complexed With Myo-Inositol
Organism: Paracoccus laeviglucosivorans nakamura 2015
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-05-23 Classification: OXIDOREDUCTASE Ligands: NAD, INS |
 |
Pyrophosphate-Dependent Kinase In The Ribokinase Family Complexed With A Pyrophosphate Analog And Myo-Inositol
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-05-16 Classification: TRANSFERASE Ligands: INS, MDN, MG, SO4 |
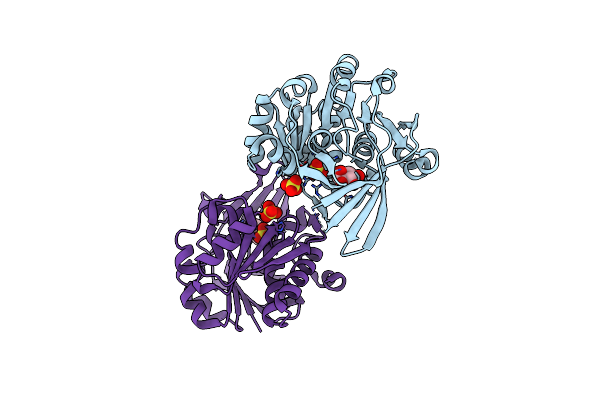 |
Sulfate-Complex Structure Of A Pyrophosphate-Dependent Kinase In The Ribokinase Family Provides Insight Into The Donor-Binding Mode
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2018-05-16 Classification: TRANSFERASE Ligands: INS, SO4 |
 |
Calcium-Dependent Phosphoinositol-Specific Phospholipase C From A Gram-Negative Bacterium, Pseudomonas Sp, Apo Form, Myoinositol Complex
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-01-18 Classification: HYDROLASE Ligands: INS, CA, PO4 |
 |
Crystal Structure Of Surfactant Protein-A Dedn Mutant (E171D/P175E/R197N/K203D) Complexed With Inositol
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-02-10 Classification: SUGAR BINDING PROTEIN Ligands: CA, INS, HEZ, CL |
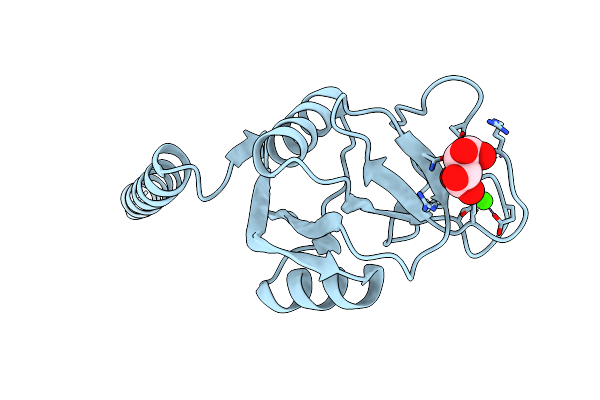 |
Crystal Structure Of Surfactant Protein-A Ded Mutant (E171D/P175E/K203D) Complexed With Inositol
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-02-10 Classification: SUGAR BINDING PROTEIN Ligands: INS, CA |
 |
Crystal Structure Of A Pentafluoro-Phe Incorporated Phosphatidylinositol-Specific Phospholipase C (H258X)From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-01 Classification: LYASE Ligands: ACT, INS |
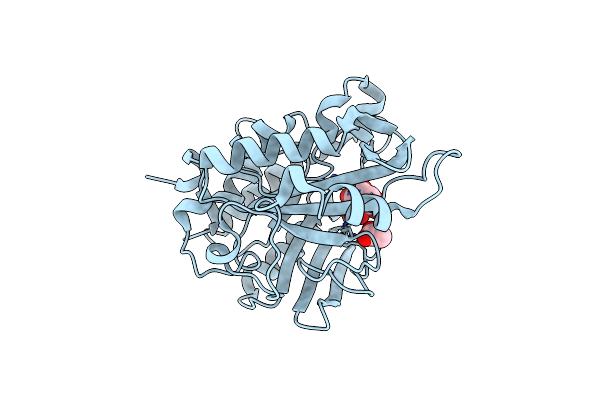 |
Structure Of The F249X Mutant Of Phosphatidylinositol-Specific Phospholipase C From Staphylococcus Aureus
Organism: Staphylococcus aureus str. newman
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-07-01 Classification: LYASE Ligands: INS, ACT |
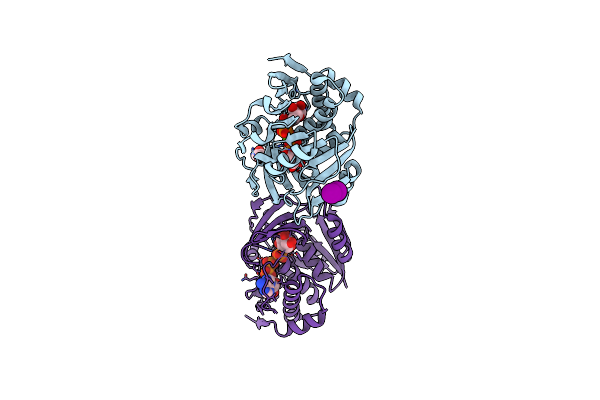 |
Myo-Inositol 3-Kinase Bound With Its Products (Adp And 1D-Myo-Inositol 3-Phosphate)
Organism: Thermococcus kodakaraensis kod1
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2015-06-03 Classification: TRANSFERASE Ligands: LIP, ADP, INS, MG, IOD, EDO |
 |
Organism: Thermococcus kodakaraensis kod1
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2015-06-03 Classification: TRANSFERASE Ligands: INS, ACP, MG, IOD |
 |
Crystal Structure Of An Abc Transporter Solute Binding Protein (Ipr025997) From Bacillus Halodurans C-125 (Bh2323, Target Efi-511484) With Bound Myo-Inositol
Organism: Bacillus halodurans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-03-25 Classification: SOLUTE BINDING PROTEIN Ligands: MG, CL, INS, NA |
 |
Crystal Structure Of Myo-Inositol Dehydrogenase From Lactobacillus Casei In Complex With Nad(H) And Myo-Inositol
Organism: Lactobacillus casei
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-03-04 Classification: OXIDOREDUCTASE Ligands: NAI, GOL, SO4, INS |
 |
Crystal Structure Of Myo-Inositol Dehydrogenase From Lactobacillus Casei In Complex With Nad And Myo-Inositol
Organism: Lactobacillus casei
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2015-03-04 Classification: OXIDOREDUCTASE Ligands: NAD, GOL, SO4, INS |
 |
Crystal Structure Of Periplasmic Abc Transporter Solute Binding Protein A7Jw62 From Mannheimia Haemolytica Phl213, Target Efi-511105, In Complex With Myo-Inositol
Organism: Mannheimia haemolytica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-12-24 Classification: TRANSPORT PROTEIN Ligands: INS, BGC, CL, ASP |
 |
Crystal Structure Of Carbohydrate Transporter Acei_1806 From Acidothermus Cellulolyticus 11B, Target Efi-510965, In Complex With Myo-Inositol
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-17 Classification: TRANSPORT PROTEIN Ligands: INS, CIT |

