Search Count: 401
 |
Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima In Complex With Zoledronate
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: ZOL, MG, LMT |
 |
Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima In Complex With Ca2+ And Etidronate
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: 911, MG, CA, LMT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-04-03 Classification: STRUCTURAL PROTEIN |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 0-60-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: LMT, PEG, PGE, PG4 |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 300-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.98 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 600-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 3600-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:4.53 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG, K, PO4 |
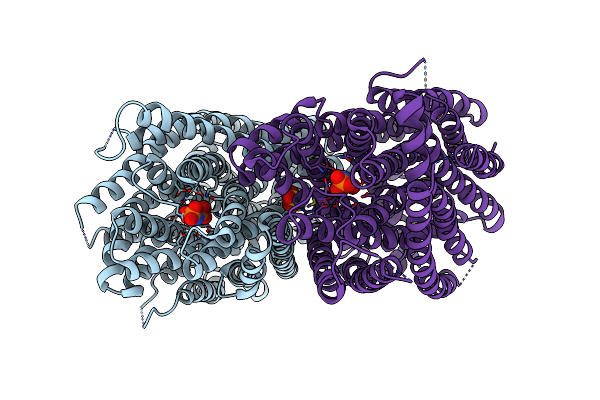 |
Crystal Structure Of Pyrobaculum Aerophilum Potassium-Independent Proton Pumping Membrane Integral Pyrophosphatase In Complex With Imidodiphosphate And Magnesium, And With Bound Sulphate
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: MG, 2PN, SO4 |
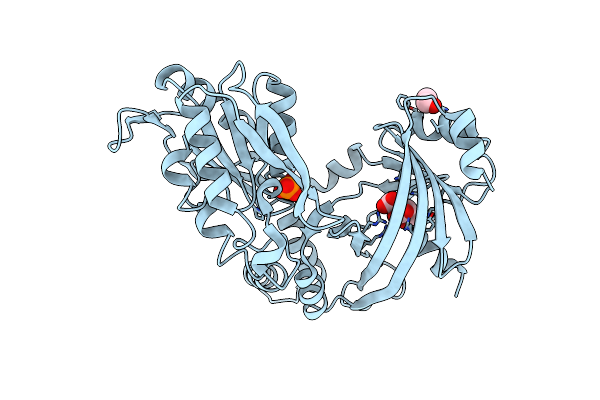 |
Crystal Structure Of The Tandem Kinase & Triphosphate Tunnel Metalloenzyme Domain Module Of The Ttm1 Protein From Arabidoposis Thaliana In Complex With Inorganic Phosphate And Citric Acid.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-04-06 Classification: UNKNOWN FUNCTION Ligands: CIT, EDO, PO4 |
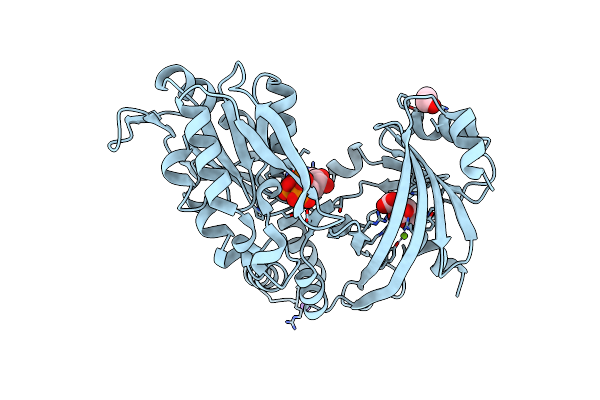 |
Crystal Structure Of The Tandem Kinase & Triphosphate Tunnel Metalloenzyme Domain Module Of The Ttm1 Protein From Arabidoposis Thaliana In Complex With A Adenosine Nucleotide Analog.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-03-23 Classification: UNKNOWN FUNCTION Ligands: CIT, EDO, ACP, MG, NA |
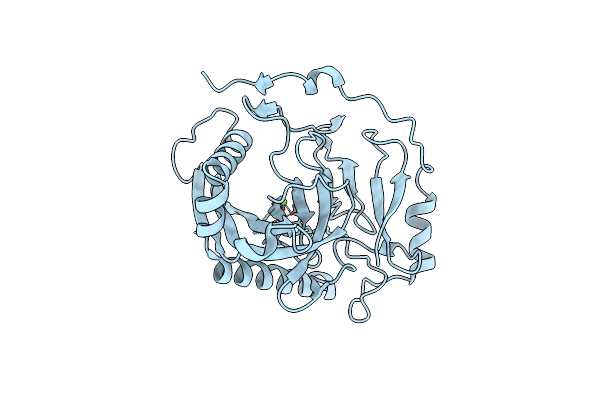 |
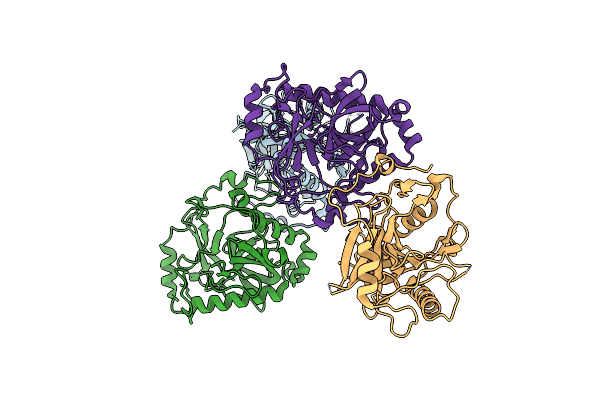
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2020-10-21 Classification: HYDROLASE Ligands: MG |
 |
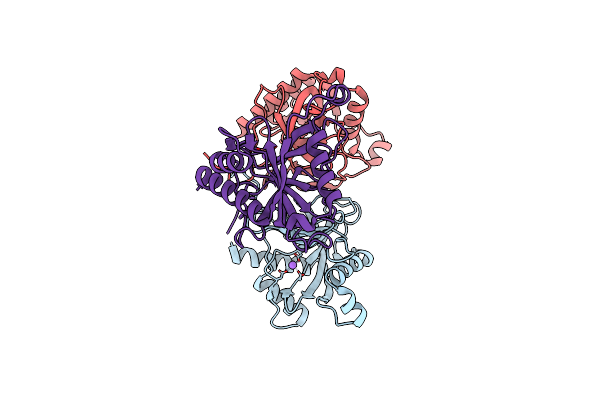
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-10-21 Classification: HYDROLASE |
 |
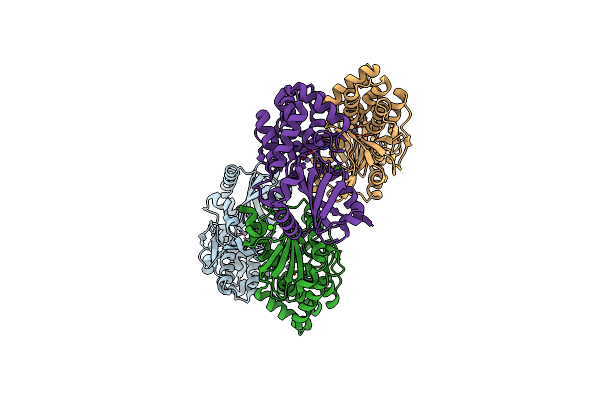
Crystal Structure Of An Inorganic Pyrophosphatase From Chlamydia Trachomatis D/Uw-3/Cx
Organism: Chlamydia trachomatis (strain d/uw-3/cx)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-04-08 Classification: HYDROLASE |
 |
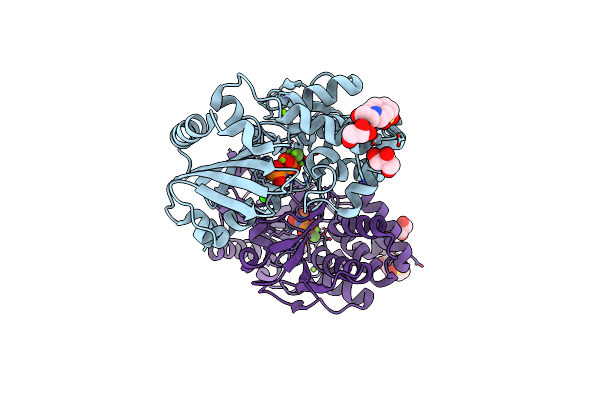
Type Ii Inorganic Pyrophosphatase (Ppase) From The Psychrophilic Bacterium Shewanella Sp. As-11, Mn-Activated Form
Organism: Shewanella sp. as-11
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-03-25 Classification: METAL BINDING PROTEIN Ligands: MN, CA |
 |
Type Ii Inorganic Pyrophosphatase (Ppase) From The Psychrophilic Bacterium Shewanella Sp. As-11, Mg-Pnp Form
Organism: Shewanella sp. as-11
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-03-25 Classification: METAL BINDING PROTEIN Ligands: MG, CA, 2PN, EPE, GOL, F |
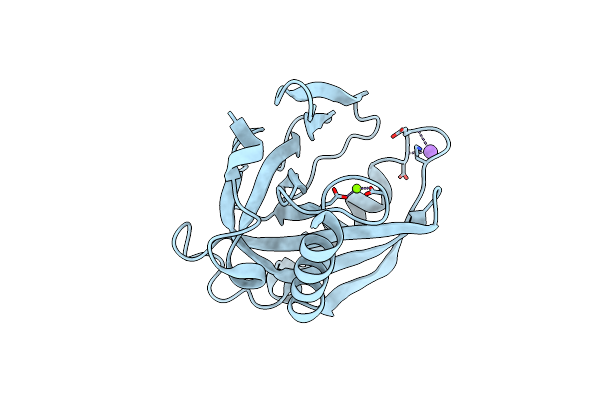 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: MG, NA |
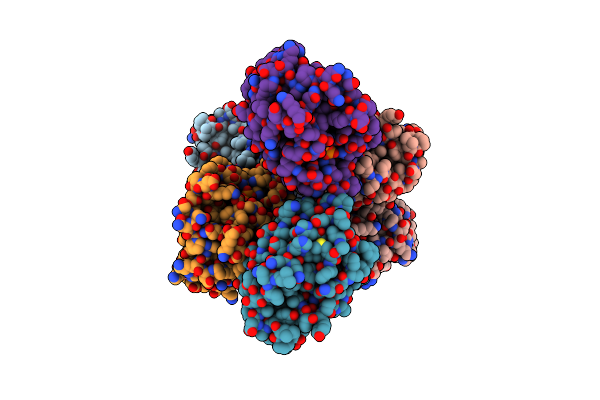 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: DPO, MG |
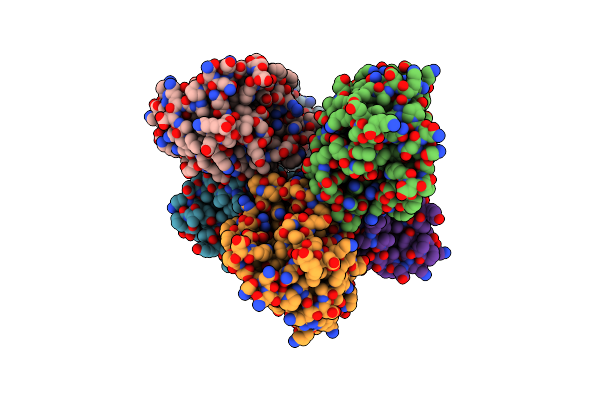 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2019-10-02 Classification: HYDROLASE |
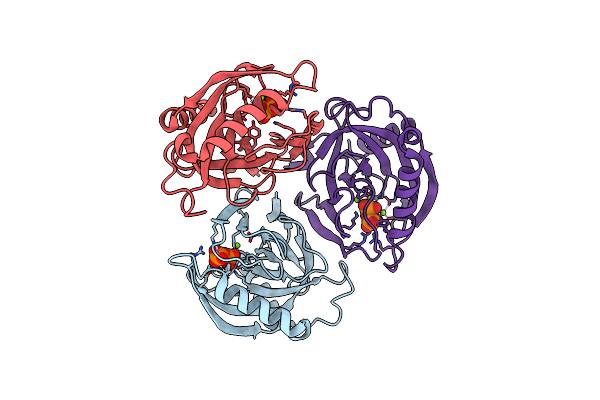 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: DPO, MG |
 |
Crystal Structure Of Inorganic Pyrophosphatase From Medicago Truncatula (R3 Crystal Form)
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-08-14 Classification: HYDROLASE |

