Search Count: 26
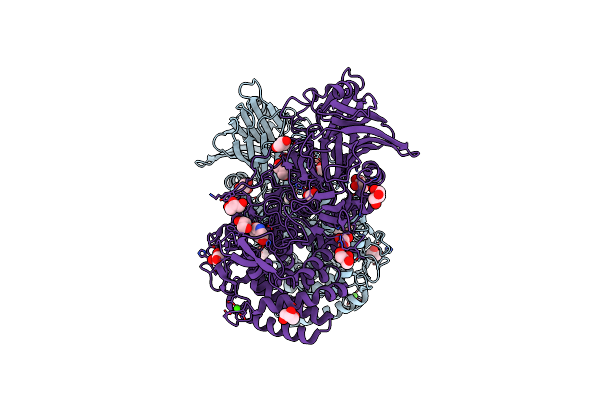 |
Crystal Structure Of Txgh116 From Thermoanaerobacterium Xylanolyticum With Isofagomine
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: IFM, CA, GOL, EDO |
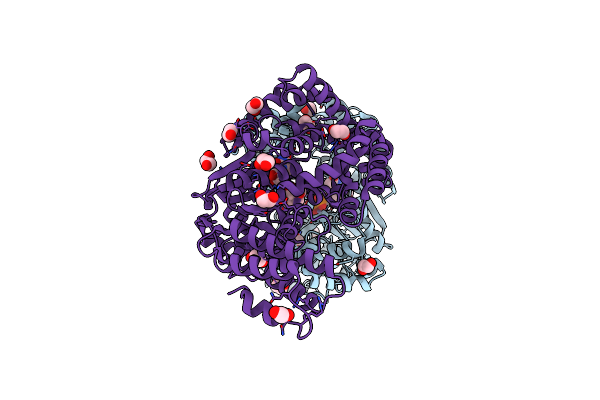 |
Crystal Structure Of Clostridium Difficile Toxin B (Tcdb) Glucosyltransferase In Complex With Udp And Isofagomine
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-11-17 Classification: TRANSFERASE/Inhibitor Ligands: EDO, UDP, IFM, MN |
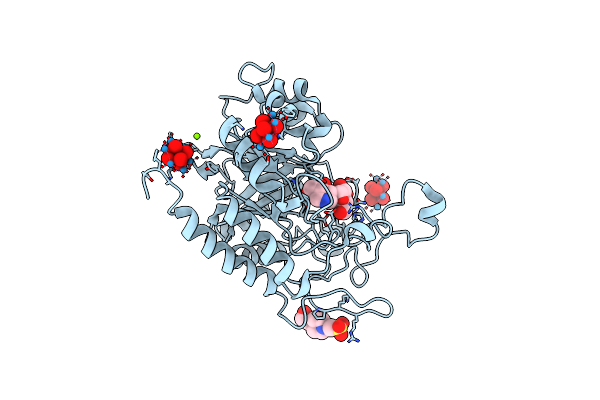 |
Structure Of The Catalytic Domain Of Human Endo-Alpha-Mannosidase Manea In Complex With Glcifg And Hexatungstotellurate(Vi) Tew
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: EPE, MG, TEW, IFM, GLC |
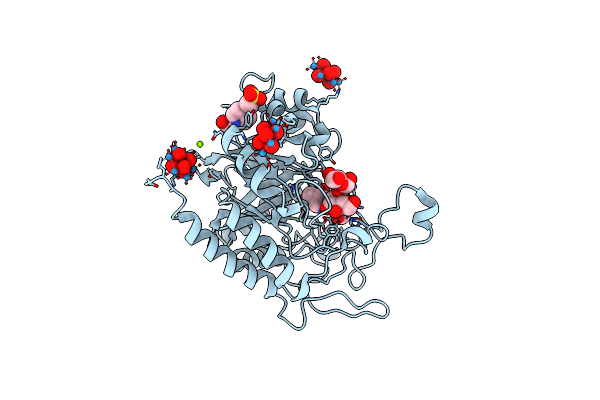 |
Structure Of The Catalytic Domain Of Human Endo-Alpha-Mannosidase Manea In Complex With Glcifg, Alpha-1,2-Mannobiose And Hexatungstotellurate(Vi) Tew
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: EPE, MG, TEW, IFM, GLC |
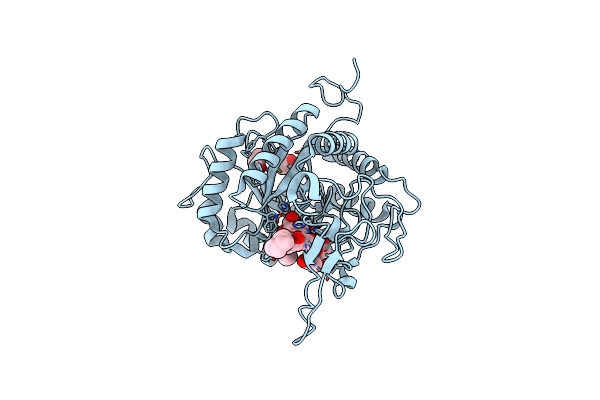 |
Structure Of The Gh99 Endo-Alpha-Mannanase From Bacteroides Xylanisolvens In Complex With Cyclohexylmethyl-Glc-1,3-Isofagomine
Organism: Bacteroides xylanisolvens xb1a
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: EDO, ACT, QTE, IFM, GLC |
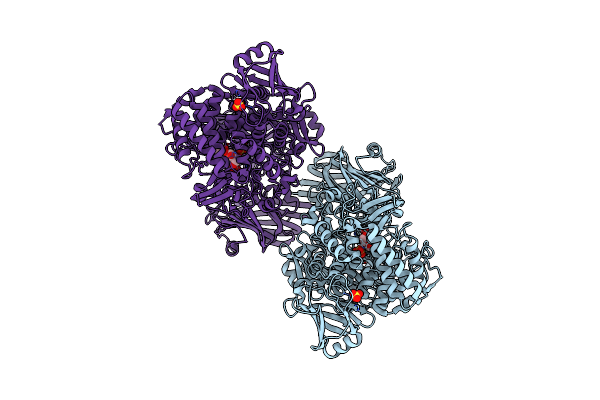 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans In Complex With Sophorose, Isofagomine, Sulfate Ion
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: IFM, SO4 |
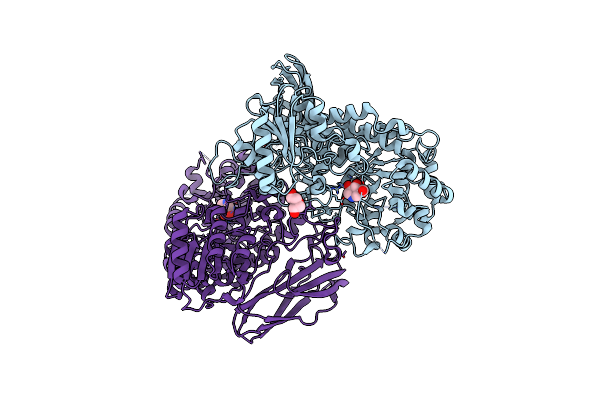 |
Crystal Structure Of Beta-Glucosidase From Listeria Innocua In Complex With Isofagomine
Organism: Listeria innocua serovar 6a (strain clip 11262)
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: IFM, MG, PEG |
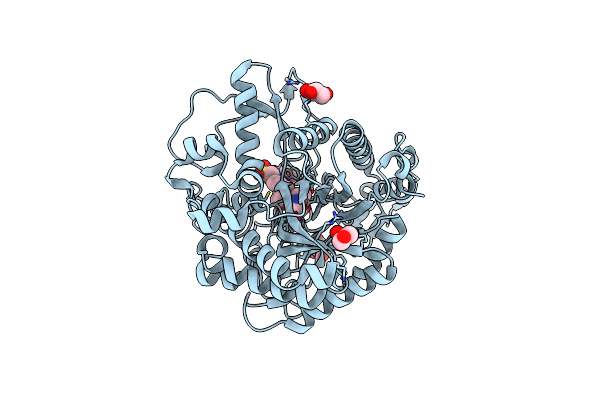 |
Different Transition State Conformations For The Hydrolysis Of Beta-Mannosides And Beta-Glucosides In The Rice Os7Bglu26 Family Gh1 Beta-Mannosidase/Beta-Glucosidase
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2015-09-16 Classification: HYDROLASE Ligands: IFM, GOL, EPE |
 |
The Catalytic Domain, Bcgh76, Of Bacillus Circulans Aman6 In Complex With 1,6-Manifg
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-03-25 Classification: HYDROLASE Ligands: IFM, MAN, EDO |
 |
Structure Of The Gh99 Endo-Alpha-Mannosidase From Bacteroides Xylanisolvens In Complex With Mannose-Alpha-1,3-Isofagomine And Alpha- 1,2-Mannobiose
Organism: Bacteroides xylanisolvens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: MAN, IFM, EDO |
 |
Structure Of The Gh99 Endo-Alpha-Mannanase From Bacteroides Xylanisolvens In Complex With Mannose-Alpha-1,3-Isofagomine
Organism: Bacteroides xylanisolvens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: MAN, IFM, EDO |
 |
Organism: Metagenomes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-09-03 Classification: HYDROLASE Ligands: NHE, IFM, NA, GOL |
 |
Crystal Structure Of 2-O-Alpha-Glucosylglycerol Phosphorylase In Complex With Isofagomine And Glycerol
Organism: Bacillus selenitireducens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-05-21 Classification: TRANSFERASE Ligands: IFM, 1PE, GOL, P6G, PGE, CA, PG4, 7PE, PE4, PE5 |
 |
The Structure Of Gh113 Beta-Mannanase Aamana From Alicyclobacillus Acidocaldarius In Complex With Manifg
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: IFM, BMA |
 |
The Structure Of Gh113 Beta-Mannanase Aamana From Alicyclobacillus Acidocaldarius In Complex With Manifg And Beta-1,4-Mannobiose
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: IFM, BMA |
 |
Crystal Structure Of Beta-Glucosidase 1 From Aspergillus Aculeatus In Complex With Isofagomine
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-04-10 Classification: HYDROLASE Ligands: NAG, MRD, MPD, NA, IFM |
 |
Structure Of The Gh99 Endo-Alpha-Mannosidase From Bacteroides Xylanisolvens In Complex With Glucose-1,3-Isofagomine
Organism: Bacteroides xylanisolvens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-02-01 Classification: HYDROLASE Ligands: GLC, IFM |
 |
Structure Of The Gh99 Endo-Alpha-Mannosidase From Bacteroides Xylanisolvens In Complex With Glucose-1,3-Isofagomine And Alpha-1,2- Mannobiose
Organism: Bacteroides xylanisolvens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-02-01 Classification: HYDROLASE Ligands: GLC, IFM |
 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase Complexed With Sulfate And Isofagomine
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-09-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: BGC, SO4, IFM, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: PO4, NAG, GOL, IFM |

