Search Count: 25
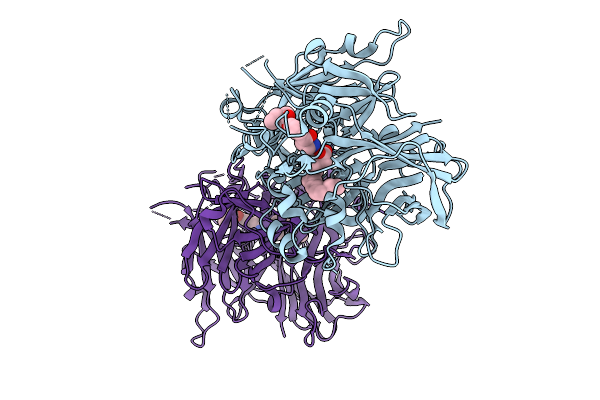 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ISOMERASE/INHIBITOR Ligands: FE2, PLM, A1BEO |
 |
Human Topoisomerase 2 Alpha Atpase Domain Bound To Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ISOMERASE Ligands: ANP, MG |
 |
Human Topoisomerase 2 Alpha Atpase Domain Bound To Bns22 And Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ISOMERASE/INHIBITOR Ligands: ANP, MG, A1ASD, CL, GOL |
 |
Human Topoisomerase 2 Beta Atpase Domain Bound To Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ISOMERASE Ligands: ANP, MG |
 |
Human Topoisomerase 2 Alpha Atpase Domain Bound To Obex 5C And Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ISOMERASE/INHIBITOR Ligands: ANP, MG, A1ASE, GOL, CL |
 |
Human Topoisomerase 2 Beta Atpase Domain Bound To Bns22 And Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ISOMERASE/INHIBITOR Ligands: ANP, A1ASD, GOL, MG |
 |
Human Topoisomerase 2 Alpha Atpase Domain Bound To Topobexin And Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ISOMERASE/INHIBITOR Ligands: ANP, MG, A1ASC, CL, GOL |
 |
Human Topoisomerase 2 Beta Atpase Domain Bound To Obex 5C And Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ISOMERASE/INHIBITOR Ligands: ANP, A1ASE, MG |
 |
Human Topoisomerase 2 Beta Atpase Domain Bound To Topobexin And Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ISOMERASE/INHIBITOR Ligands: ANP, A1ASC, MG |
 |
Crystal Structure Of L-Ribulose 3-Epimerase From Arthrobacter Globiformis M30
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: MG, PEG |
 |
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: PSJ, FUD, MG, NA, PEG |
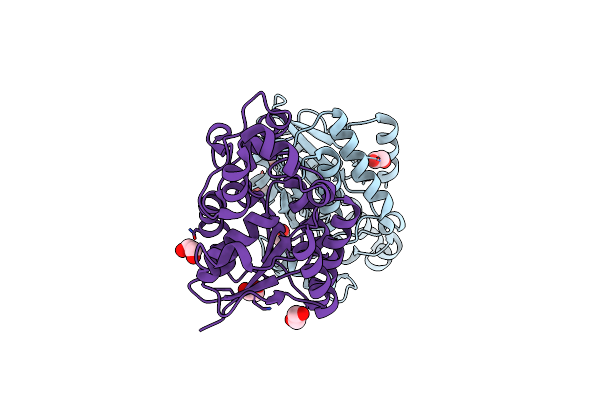 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-22 Classification: ISOMERASE Ligands: EDO |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2023-05-10 Classification: Hydrolase, Isomerase Ligands: 12P, ZN, CL |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2023-05-10 Classification: Hydrolase, Isomerase Ligands: ZN |
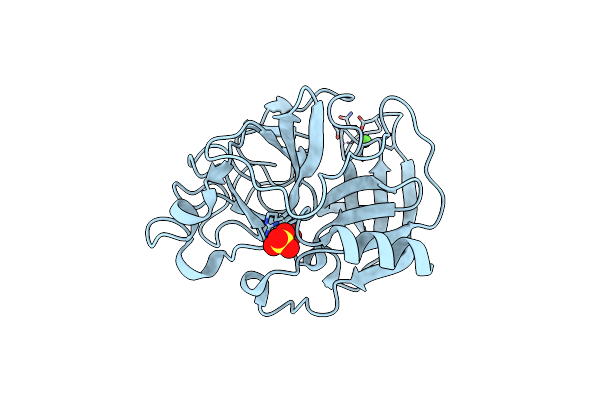 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-07-06 Classification: ISOMERASE Ligands: CA, SO4, SO3 |
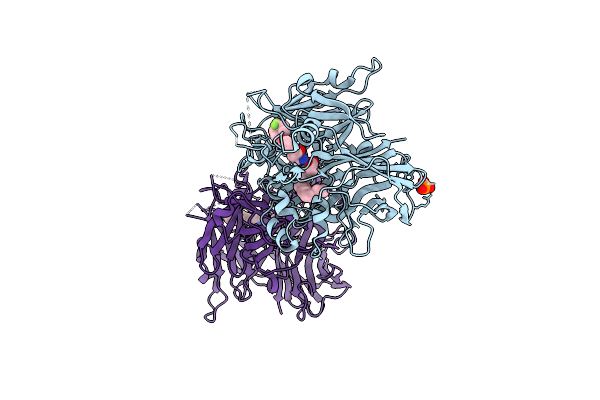 |
Crystal Structure Of Bovine Rpe65 In Complex With Gem-Difluoro Emixustat And Palmitate
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-06-02 Classification: HYDROLASE/INHIBITOR Ligands: FE2, PLM, XQ7, PO4 |
 |
Crystal Structure Of Udp-Glucose 4-Epimerase From Bifidobacterium Longum In Complex With Nad+ And Udp-Glcnac
Organism: Bifidobacterium longum subsp. longum (strain atcc 15707 / dsm 20219 / jcm 1217 / nctc 11818 / e194b)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-08-07 Classification: ISOMERASE Ligands: PEG, NAD, UD1, MG |
 |
Crystal Structure Of Udp-Glucose 4-Epimerase From Bifidobacterium Longum In Complex With Nad+ And Udp-Glc
Organism: Bifidobacterium longum subsp. longum (strain atcc 15707 / dsm 20219 / jcm 1217 / nctc 11818 / e194b)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-08-07 Classification: ISOMERASE Ligands: NAD, UPG |
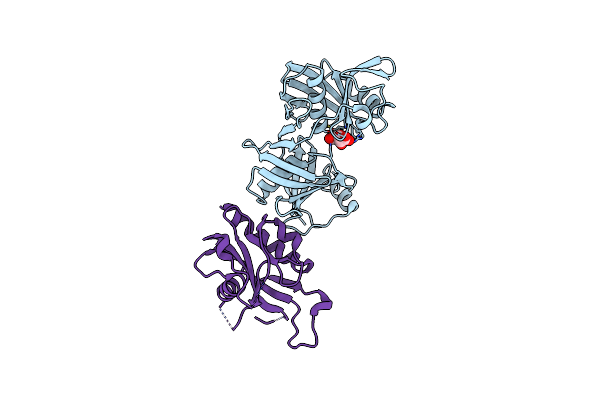 |
Crystal Structure Of Escherichia Coli (Strain K12) Mrna Decapping Complex Rpph-Dapf
Organism: Escherichia coli (strain k12), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-06-06 Classification: ISOMERASE/HYDROLASE Ligands: TLA, IOD |
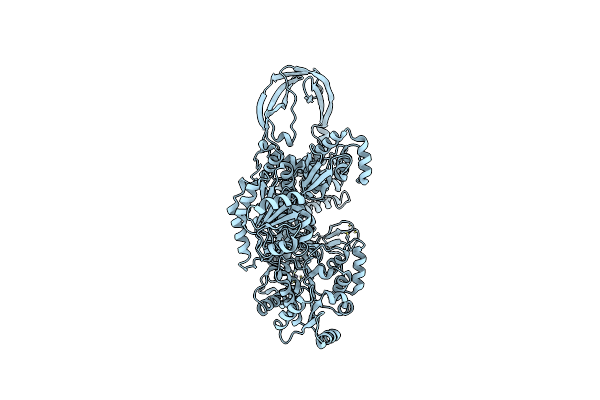 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-12-26 Classification: ISOMERASE Ligands: ZN |

