Search Count: 712
 |
Organism: Ebola virus - mayinga, zaire, 1976
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: VIRAL PROTEIN |
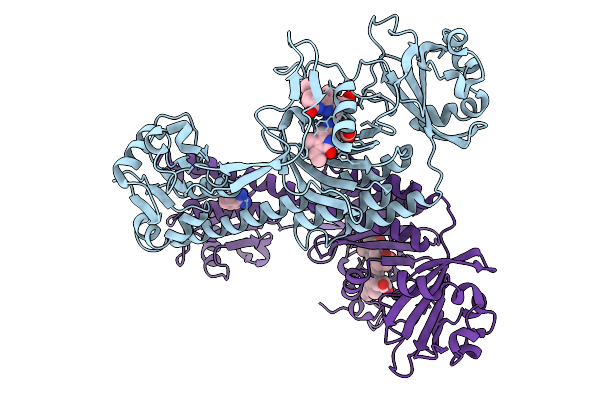 |
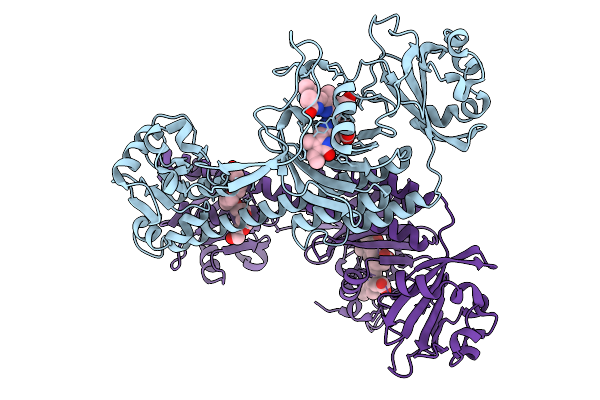
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
1.8 A Resolution Structure Of The Photosystem I Assembly Intermediate Lacking Stromal Subunits.
Organism: Synechocystis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: PHOTOSYNTHESIS Ligands: LHG, CL0, CLA, PQN, BCR, ECH, LMG, LMT, CL, SF4, 5X6 |
 |
Hiv-1 Envelope Glycoprotein (Bg505 Gp140 Sosip.664) Trimer In Complex With Three Copies Of Elc07 Broadly Neutralizing Antibody.
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: VIRAL PROTEIN Ligands: NAG |
 |
X-Ray Structure Of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2025-04-16 Classification: DE NOVO PROTEIN Ligands: CU |
 |
X-Ray Structure Of A Second-Sphere H-Bond Deletion Mutant Of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-04-16 Classification: DE NOVO PROTEIN Ligands: CU |
 |
Hiv-1 Envelope Glycoprotein (Bg505 Gp140 Sosip.664) Trimer In Complex With Elc07 Broadly Neutralizing Antibody.
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure Of The Sars-Cov-2 S 6P Trimer Complex With The Human Neutralizing Antibody Fab Fragment, C1533 (Local Refinement Of Ntd And C1533)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structure Of The Sars-Cov-2 S 6P Trimer Complex With The Human Neutralizing Antibody Fab Fragment, C1596
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structure Of The Sars-Cov-2 S 6P Trimer Complex With The Human Neutralizing Antibody Fab Fragment, C952
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mn And Amp-Pnp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ANP, MN |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mg And Amp-Pnp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ANP, MG |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Apo)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-06 Classification: LIGASE |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mg And Adp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ADP, MG, PG4 |
 |
Organism: Streptomyces tsukubensis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-10-09 Classification: LIGASE Ligands: ATP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-07-10 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, CL, GOL, YIL |

