Search Count: 28
 |
Solution Structure Of Staphylococcus Aureus Response Regulator Arlr Dna-Binding Domain
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: SOLUTION NMR Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DNA BINDING PROTEIN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-08-30 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: TRANSPORT PROTEIN |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-31 Classification: CARBOHYDRATE Ligands: GOL, ZN, CA |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-05-31 Classification: CARBOHYDRATE Ligands: BCT, ZN, CA |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-06-23 Classification: SIGNALING PROTEIN |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-06-23 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
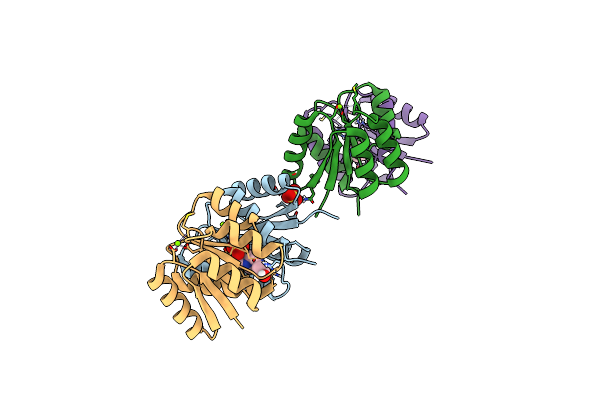
Crystal Structure Of Staphylococcus Aureus Response Regulator Arlr Receiver Domain In Complex With Bef3 And Mg
Organism: Staphylococcus aureus (strain bovine rf122 / et3-1)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2019-10-30 Classification: SIGNALING PROTEIN Ligands: BEF, MG, IMD |
 |
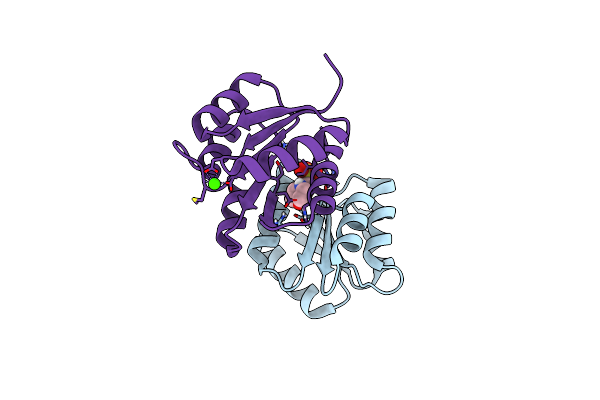
Crystal Structure Of Staphylococcus Aureus Response Regulator Arlr Receiver Domain In Complex With Mg
Organism: Staphylococcus aureus (strain bovine rf122 / et3-1)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2019-10-30 Classification: SIGNALING PROTEIN Ligands: MG, SO4, EPE, IMD |
 |
Crystal Structure Of Staphylococcus Aureus Response Regulator Arlr Receiver Domain
Organism: Staphylococcus aureus rf122
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-10-30 Classification: SIGNALING PROTEIN Ligands: CA, MES |
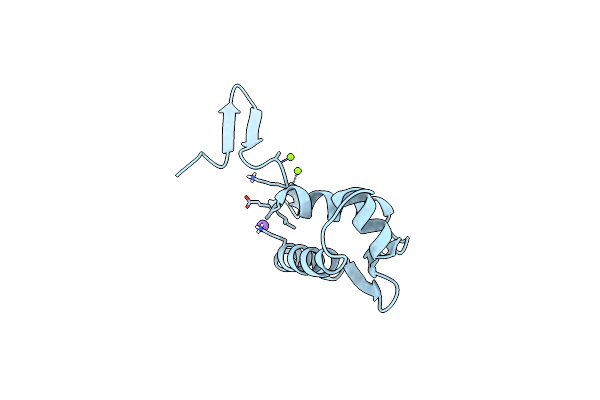 |
Crystal Structure Of Staphylococcus Aureus Response Regulator Arlr Dna Binding Domain
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-10-30 Classification: DNA BINDING PROTEIN Ligands: MG, NA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2019-08-14 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-11-14 Classification: METAL BINDING PROTEIN Ligands: CA, WW7 |
 |
Nmr Structure Of The N-Domain Of Troponin C Bound To Switch Region Of Troponin I And 3-Methyldiphenylamine (Solvent Exposed Mode)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2017-07-19 Classification: METAL BINDING PROTEIN Ligands: 9XG |
 |
Nmr Structure Of The N-Domain Of Troponin C Bound To Switch Region Of Troponin I And 3-Methyldiphenylamine (Peptide Mode)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2017-07-12 Classification: METAL BINDING PROTEIN Ligands: 9XG |
 |
Nmr Structure Of The N-Domain Of Troponin C Bound To Switch Region Of Troponin I
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2017-05-24 Classification: METAL BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-08-10 Classification: TRANSFERASE Ligands: ACT, LDA, MPD |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2002-09-13 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2002-09-13 Classification: TRANSFERASE |

