Search Count: 144
 |
Organism: Clostridium phage phicd111
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIBIOTIC Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: A1A3I |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: A1A3J |
 |
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: RNA Ligands: CA, K, 5GP |
 |
Cryoem Structure Of The Candida Albicans Group I Intron-Compound 11 Complex Under Magnesium Condition
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: RNA Ligands: A1BNU, MG, K |
 |
Cryoem Structure Of The Candida Albicans Group I Intron-Compound 11 Complex Under Calcium Condition
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: RNA Ligands: A1BNU, K, CA |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-16 Classification: PROTEIN BINDING |
 |
Organism: Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
 |
Organism: Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
 |
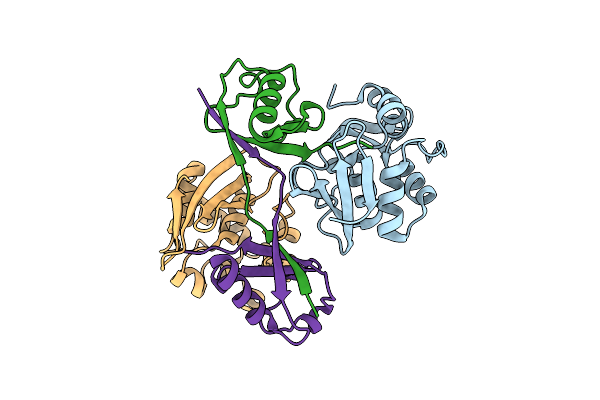
Structure Of A Dimeric Ubiquitin Variant (Ubv3) That Inhibits The Protease (Pro) From Turnip Yellow Mosaic Virus (Tymv)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2025-01-22 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Turnip Yellow Mosaic Virus (Tymv) Protease (Pro) Bound To A Ubiquitin Variant (Ubv3)
Organism: Turnip yellow mosaic virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Duf2436 Domain Which Is Frequently Found In Virulence Proteins From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis w83
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2024-10-09 Classification: UNKNOWN FUNCTION |
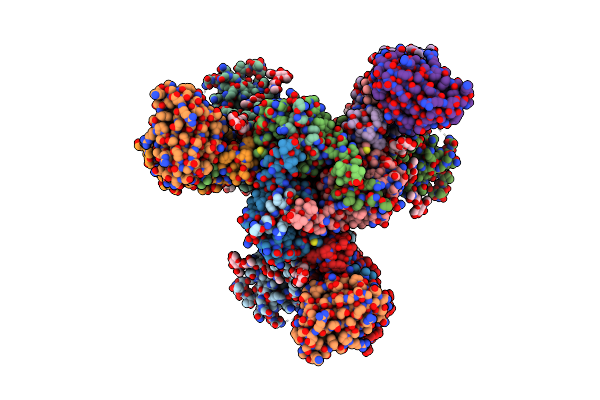 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-B.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
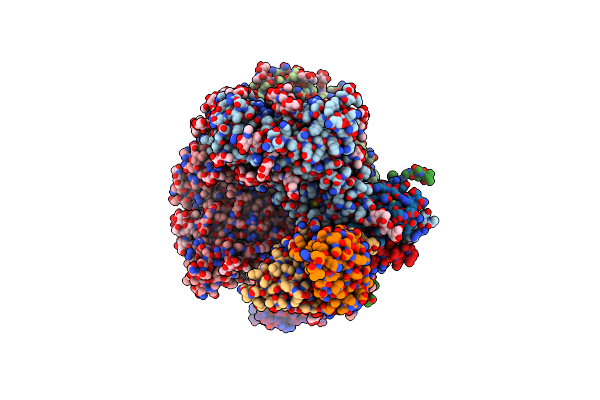 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-C.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
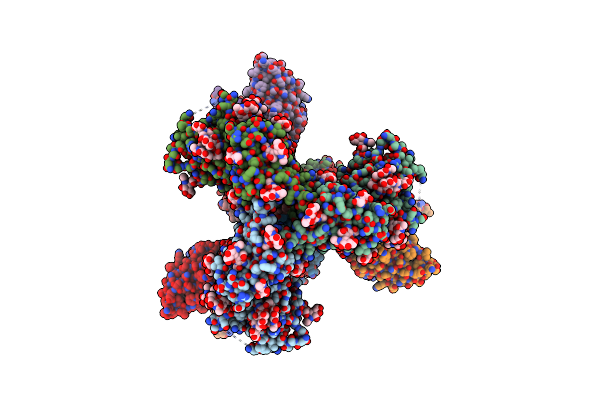 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-D.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
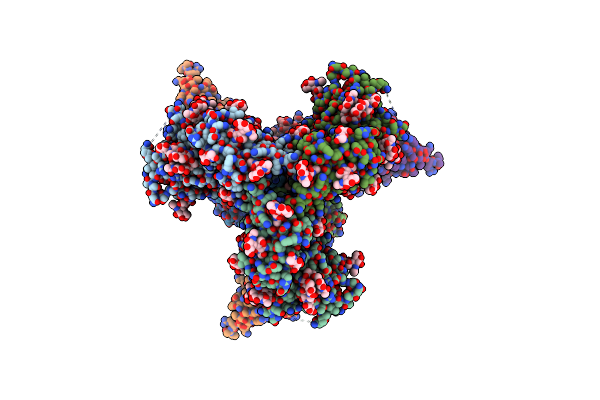 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-E.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
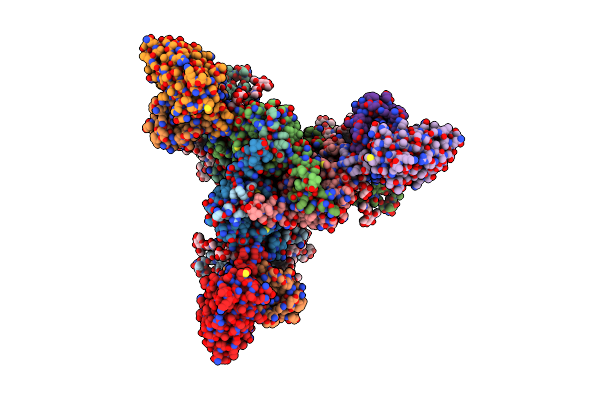 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Herh-C.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
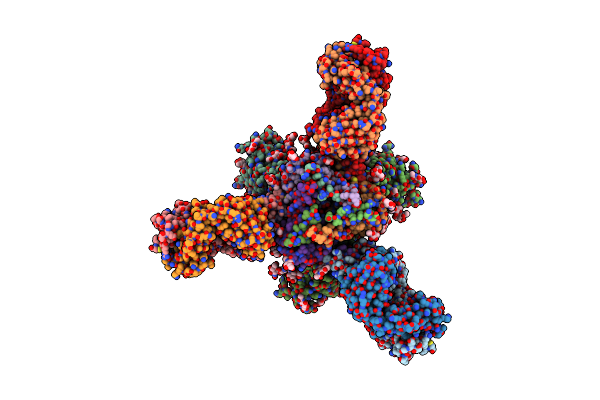 |
Cryo-Em Structure Of Trnm-B*01 Fab In Complex With Hiv-1 Env Trimer Bg505.Ds Sosip
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
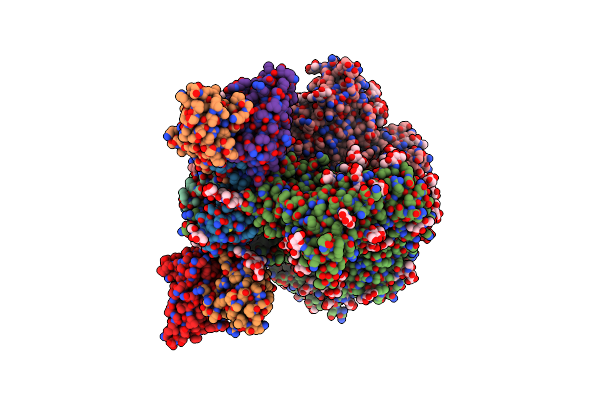 |
Cryo-Em Structure Of Herh-B*01 Fab In Complex With Hiv-1 Env Trimer Bg505.Ds Sosip
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |

