Search Count: 14
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-03-29 Classification: GENE REGULATION Ligands: U59, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-03-29 Classification: GENE REGULATION/INHIBITOR Ligands: U7R, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-03-29 Classification: GENE REGULATION/INHIBITOR Ligands: UHI, EDO |
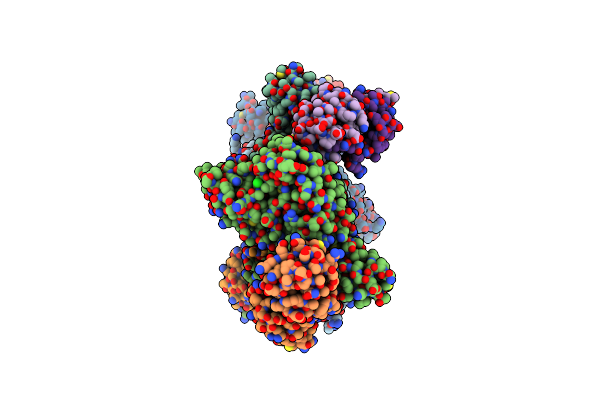 |
Structure Of The Sars-Cov2 Plpro (C111S) In Complex With A Dimeric Ubv That Inhibits Activity By An Unusual Allosteric Mechanism
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-01-25 Classification: VIRAL PROTEIN/SIGNALING PROTEIN Ligands: ZN, BR, NA, CL |
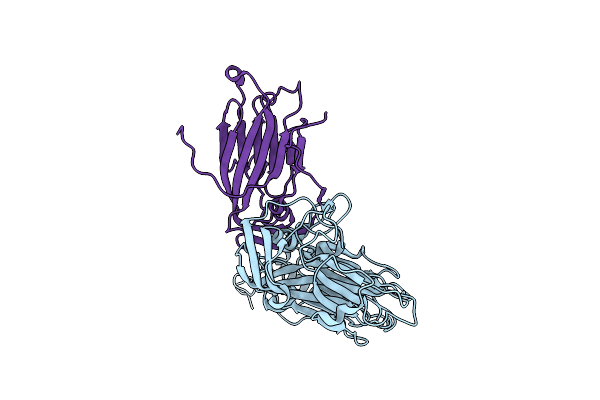 |
Crystal Structure And Proteomics Analysis Of Empty Virus Like Particles Of Cowpea Mosaic Virus
Organism: Cowpea mosaic virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-03-09 Classification: VIRUS |
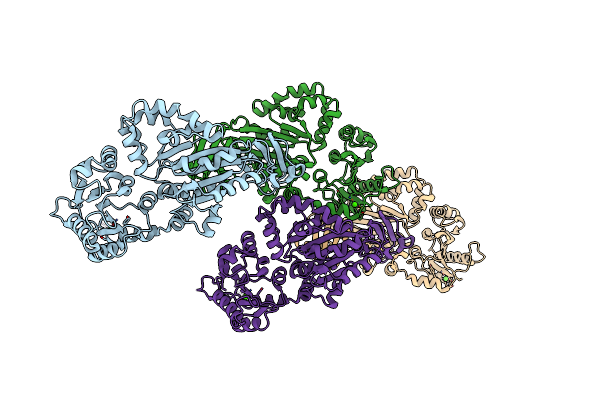 |
Organism: Photobacterium damselae
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2014-12-03 Classification: TRANSFERASE Ligands: CA |
 |
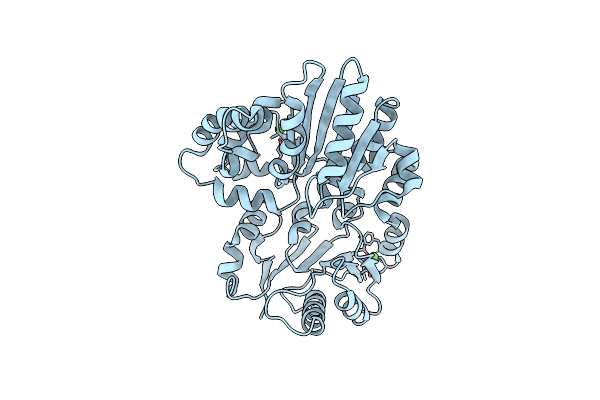
Crystal Structure Of Sialyltransferase From Photobacterium Damsela With Cmp-3F(A)Neu5Ac Bound
Organism: Photobacterium damselae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-12-03 Classification: TRANSFERASE Ligands: CSF, CA |
 |
Crystal Structure Of Sialyltransferase From Photobacterium Damselae, Residues 113-497 Corresponding To The Gt-B Domain
Organism: Photobacterium damselae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-03 Classification: TRANSFERASE Ligands: CA |
 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-11-06 Classification: LYASE Ligands: PO4, EDO |
 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase Trapped With Pyruvate Covalently Bound Through A Schiff Base To Lys164
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-11-06 Classification: LYASE Ligands: PO4, EDO |
 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase K164A Mutant
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-11-06 Classification: LYASE Ligands: ME2, PG6 |
 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase K164 Mutant Complexed With N-Acetylneuraminic Acid
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-11-06 Classification: LYASE Ligands: SI3, ME2, CL, IPA |
 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase K164 Mutant Complexed With N-Glycolylneuraminic Acid
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-11-06 Classification: LYASE Ligands: NGF, ME2 |
 |
Crystal Structure Of Sr Protein Kinase 1 Complexed To Its Substrate Asf/Sf2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-04-01 Classification: Transferase/Splicing Ligands: SEP, ALA, ANP |

