Search Count: 19
 |
Erap1 In Complex With 1-[2-(3-Oxo-3,4-Dihydro-2H-1,4-Benzothiazin-4-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, B3P, EDO, BR, A1IMK |
 |
Erap1 In Complex With 1-[2-(6-Bromo-3-Oxo-3,4-Dihydro-2H-1,4-Benzoxazin-4-Yl)Acetamido]-4,4-Difluorocyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, PO4, EDO, A1IMJ |
 |
Erap1 In Complex With 1-[2-(6-Chloro-3-Oxo-3,4-Dihydro-2H-1,4-Benzothiazin-4-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, MLT, EDO, A1IMM |
 |
Erap1 In Complex With 1-[2-(2-Oxo-5-Phenyl-2,3-Dihydro-1,3-Benzothiazol-3-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, PO4, EDO, A1IML |
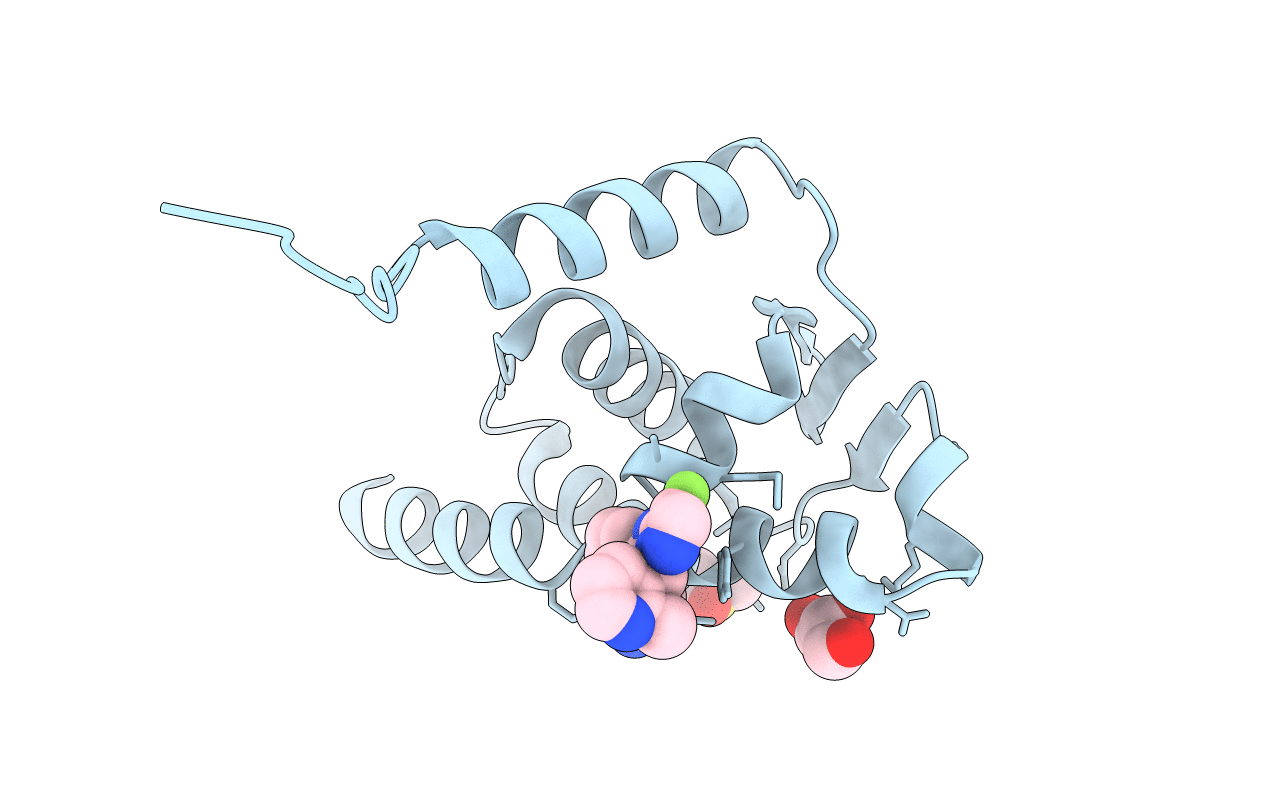 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-07-28 Classification: TRANSCRIPTION Ligands: TEQ, GOL, DMS |
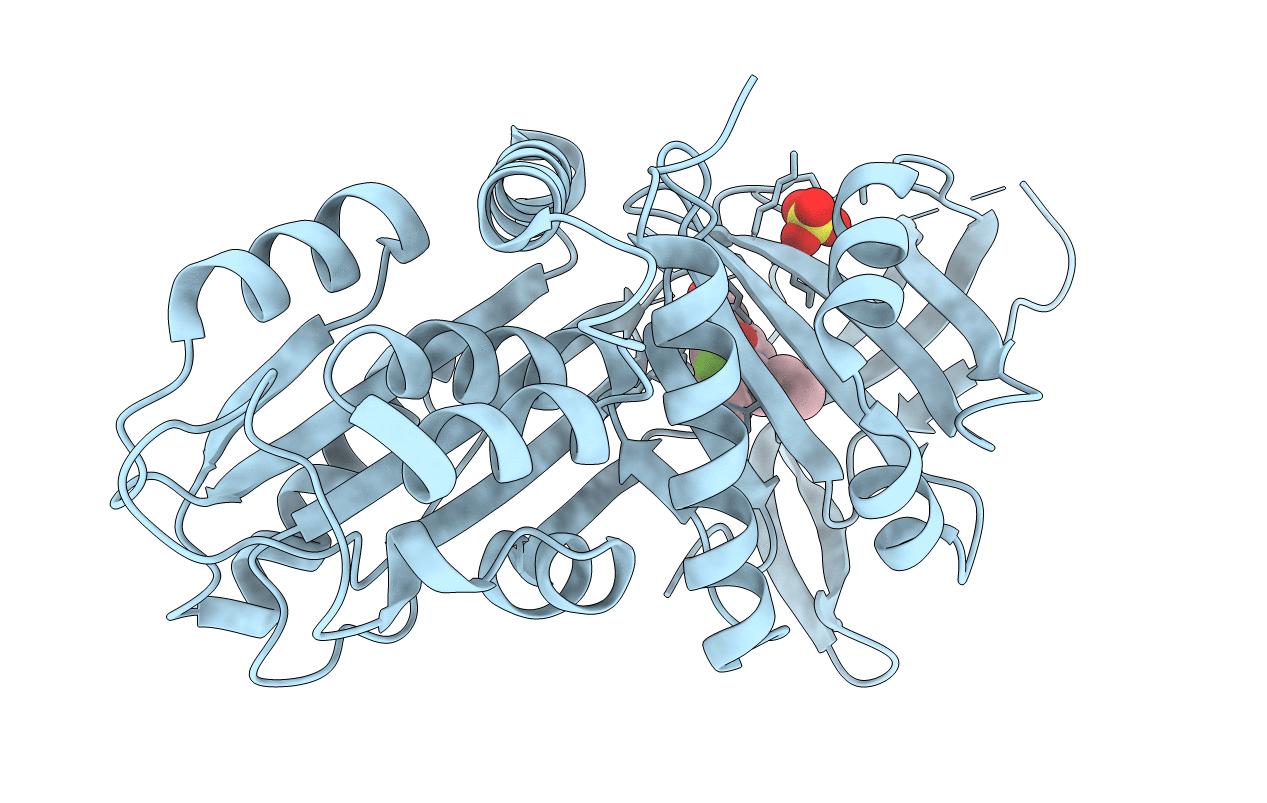 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-03-10 Classification: PROTEIN BINDING Ligands: R7Z, SO4 |
 |
Human Endoplasmic Reticulum Aminopeptidase 1 (Erap1) In Complex With (4Ar,5S,6R,8S,8Ar)-5-(2-(Furan-3-Yl)Ethyl)-8-Hydroxy-5,6,8A-Trimethyl-3,4,4A,5,6,7,8,8A-Octahydronaphthalene-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2020-03-18 Classification: HYDROLASE Ligands: ZN, MLT, EDO, MNZ |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-05-30 Classification: TRANSFERASE Ligands: NI, AGS, MG |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, GOL |
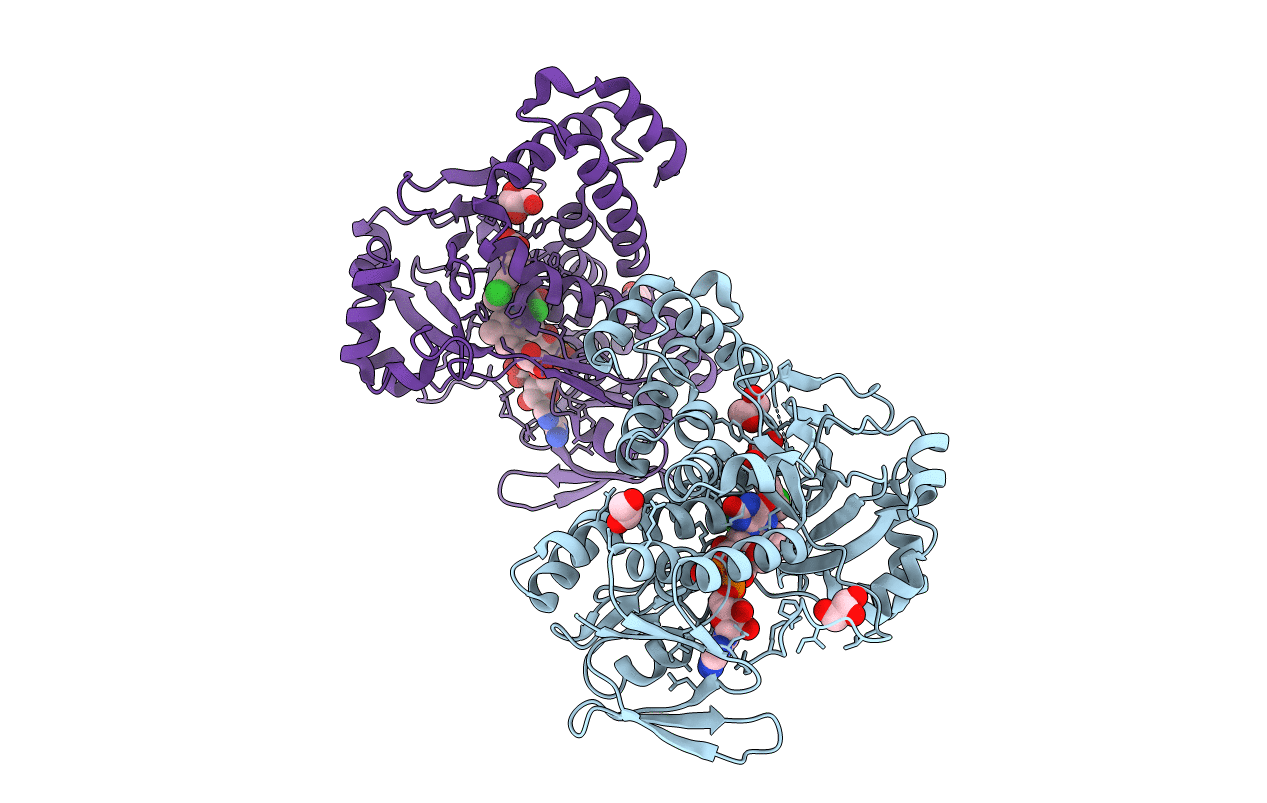 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5-Chloro-6-Methyl-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, 8RK, GOL |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-2-Oxo-6-[(1R)-1-(Pyridin-2-Yl)Ethoxy]-2,3-Dihydro-1,3-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8R8 |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-6-[(1R)-1-(Pyridin-2-Yl)Ethoxy]-1,2-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8R5 |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-6-[(1R)-1-(6-Methylpyridazin-3-Yl)Ethoxy]-1,2-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8RB |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With The Enzyme Substrate L-Kynurenine
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, GOL, KYN |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5,6-Dichloro-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2017-06-14 Classification: OXIDOREDUCTASE Ligands: FAD, CL, JHY, GOL |
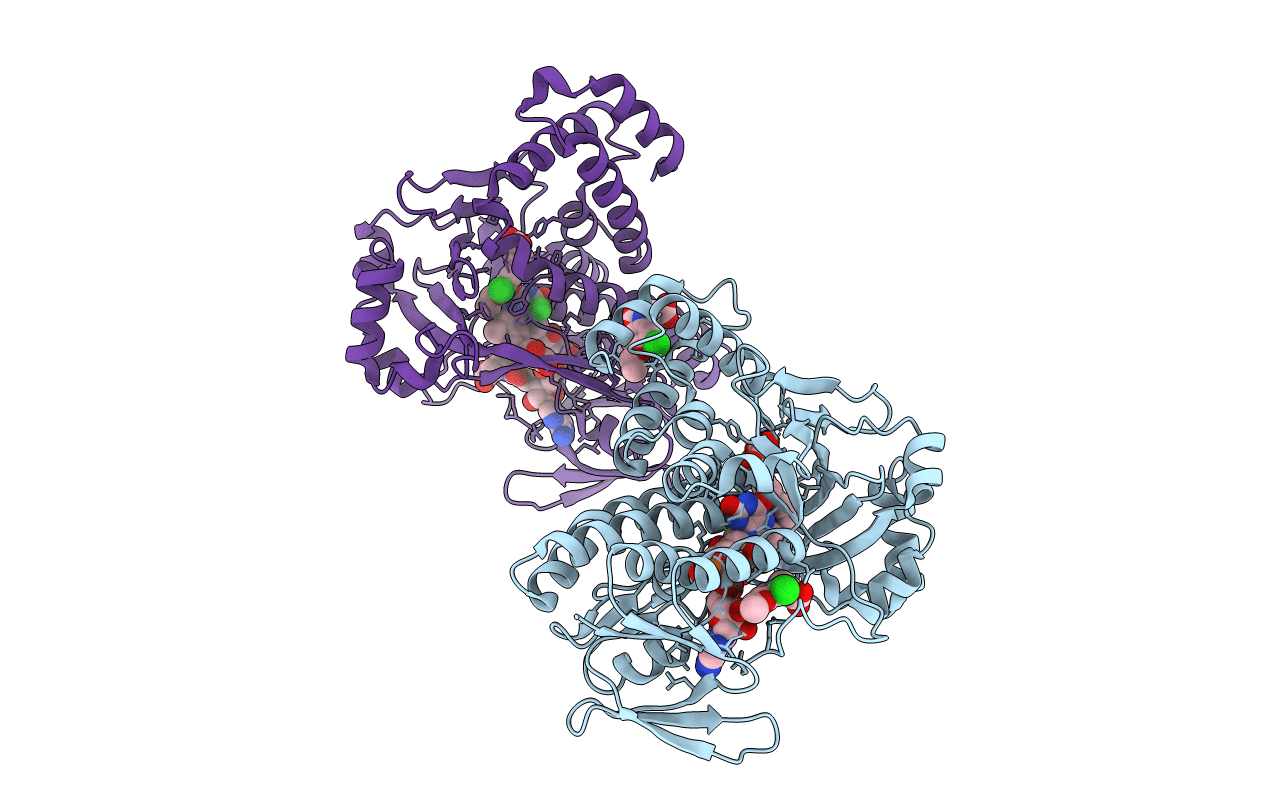 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5-Chloro-6-Ethoxy-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-04-19 Classification: OXIDOREDUCTASE Ligands: FAD, CL, 8EQ, GOL |
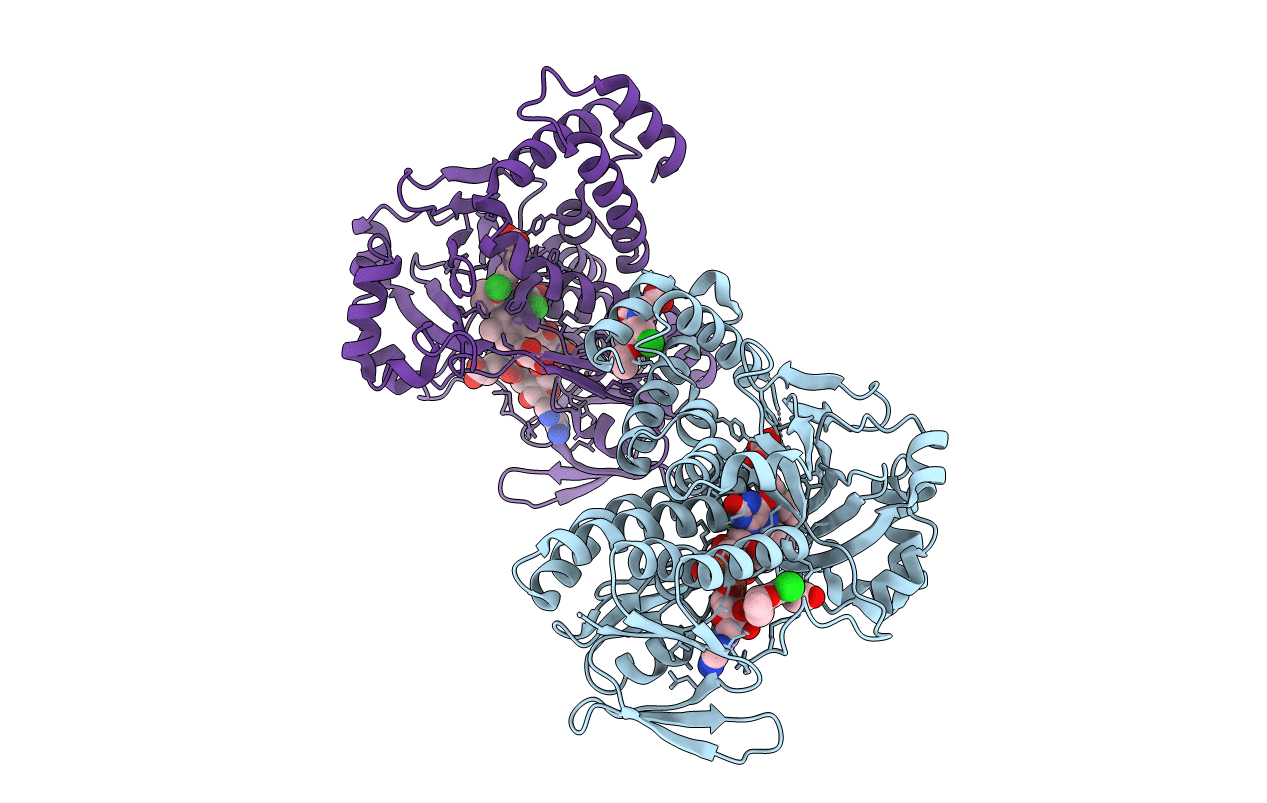 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5-Chloro-6-Cyclopropoxy-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2017-04-19 Classification: OXIDOREDUCTASE Ligands: FAD, CL, FYK, GOL |
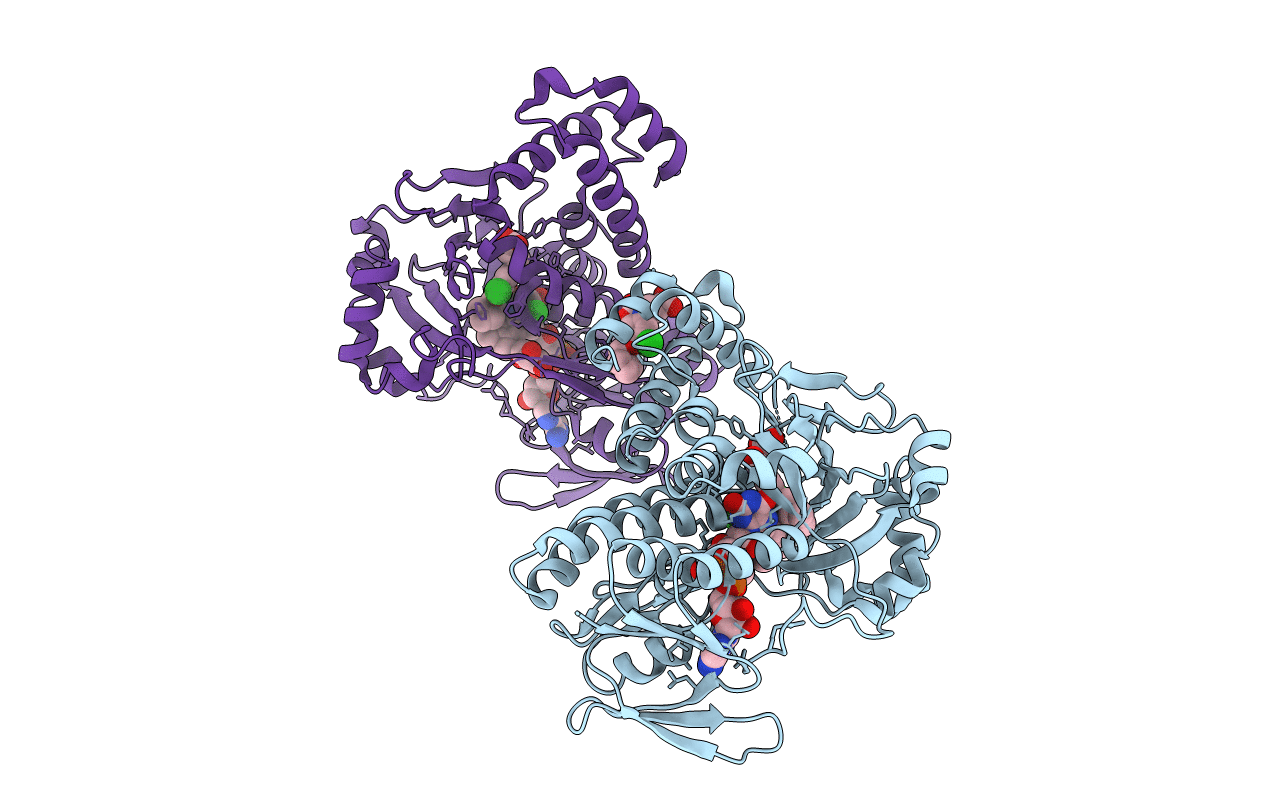 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-[5-Chloro-6-(Cyclobutylmethoxy)-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl]Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-04-19 Classification: OXIDOREDUCTASE Ligands: FAD, CL, OK1, GOL |
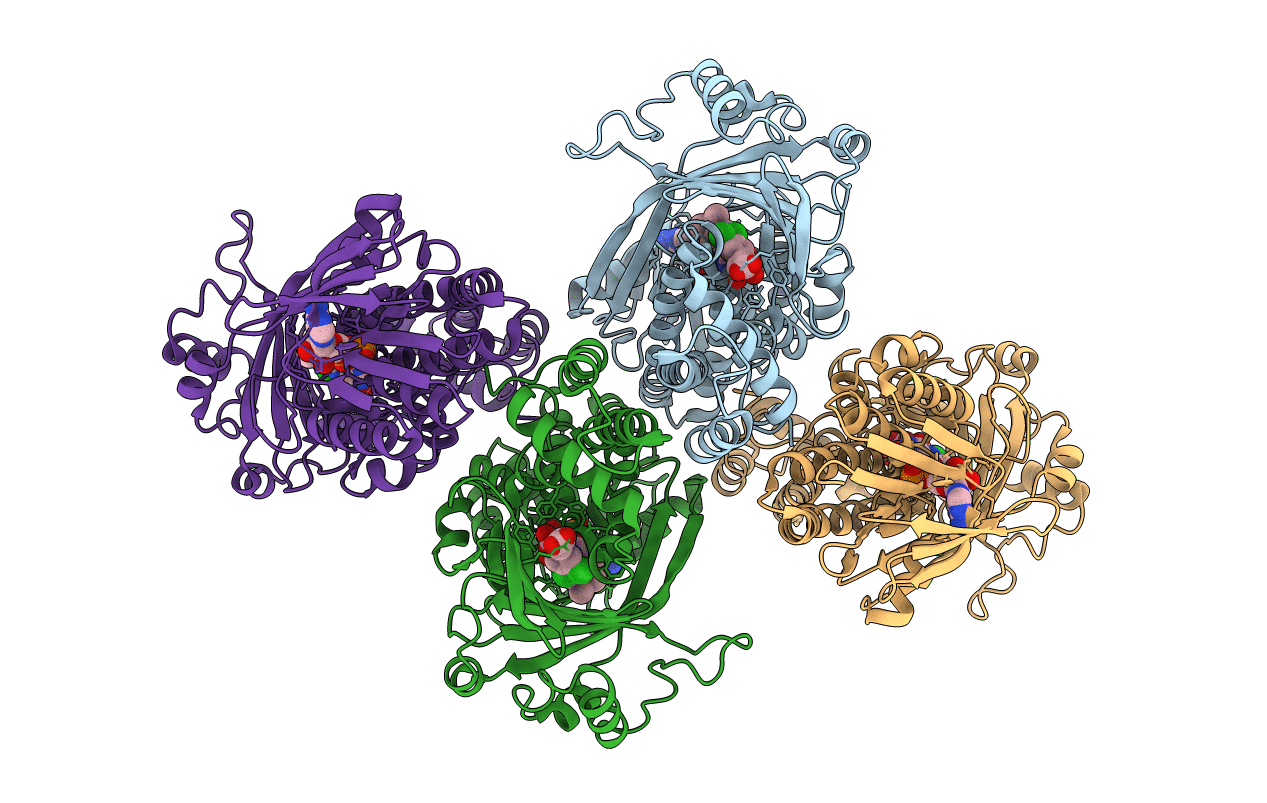 |
Crystal Structure Of Pseudomonas Fluorescens Kynurenine-3- Monooxygenase (Kmo) In Complex With Gsk180
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2016-01-13 Classification: OXIDOREDUCTASE Ligands: FAD, JHY |

