Search Count: 85
 |
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PEPTIDE BINDING PROTEIN Ligands: TEW, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: HYDROLASE/PROTEIN BINDING Ligands: ADP, MG, ATP, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: HYDROLASE/PROTEIN BINDING Ligands: MG, ATP, ADP, ZN |
 |
Rpn1/Nub1Ubl-Focused Alignment Of The Non-Substrate-Engaged Human 26S Proteasome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: HYDROLASE/PROTEIN BINDING |
 |
Organism: Staphylococcus aureus subsp. aureus mu50, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-06-12 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Bifunctional Inhibition Of Neutrophil Elastase And Cathepsin G By Eap3 Of S. Aureus
Organism: Staphylococcus aureus subsp. aureus mu50, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-06-12 Classification: PROTEIN BINDING Ligands: NAG |
 |
Bifunctional Inhibition Of Neutrophil Elastase And Cathepsin G By Eap4 Of S. Aureus
Organism: Staphylococcus aureus subsp. aureus mu50, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-06-12 Classification: PROTEIN BINDING Ligands: NAG |
 |
Organism: Staphylococcus aureus subsp. aureus mu50, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-06-12 Classification: PROTEIN BINDING |
 |
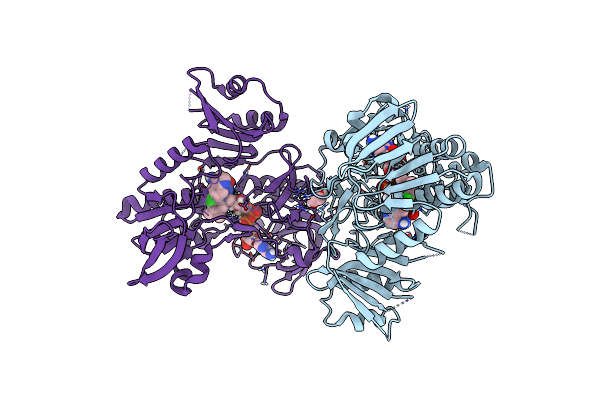
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 6-Fluoroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QDI, EDO, CL |
 |
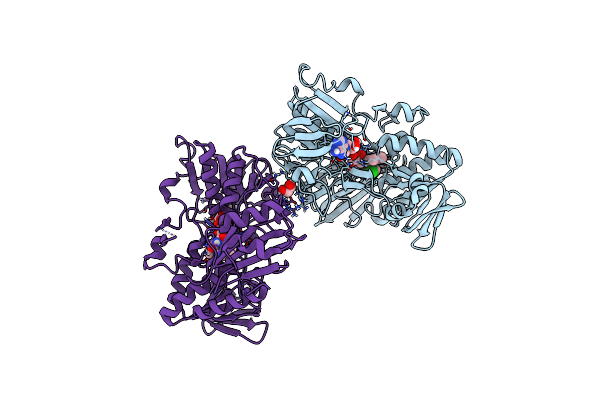
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 6-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QD1, EDO |
 |
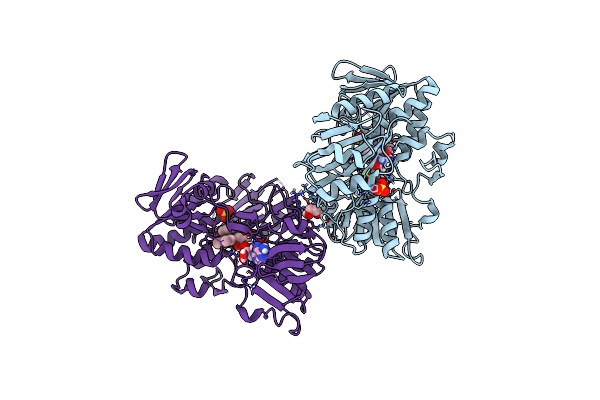
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 7-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QBC, EDO |
 |
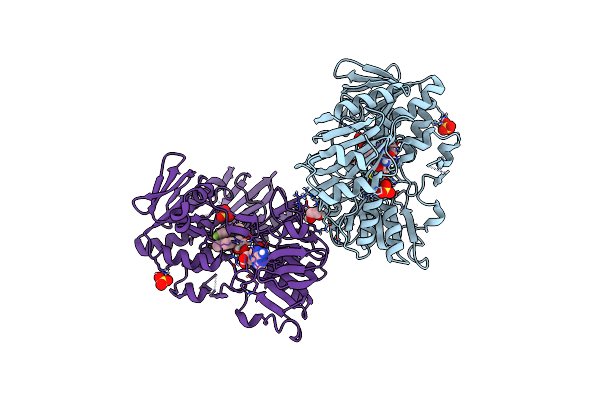
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With Quinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: 1LM, EDO, FAD, SO4 |
 |
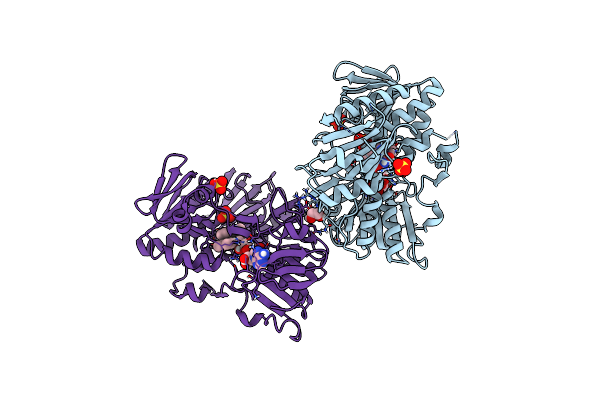
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 6-Fluoro-2-Methylquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: EDO, FAD, QEF, SO4 |
 |
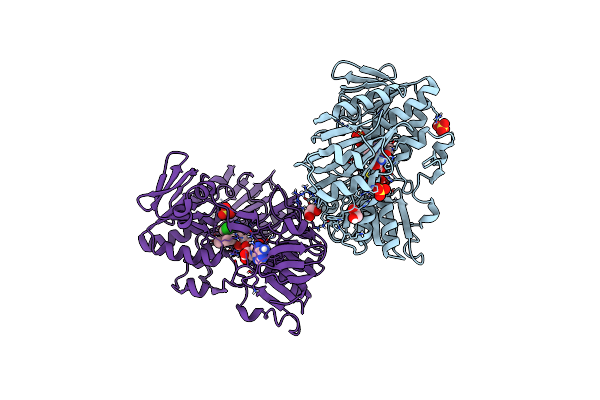
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 8-Fluoro-2-Methylquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QC6, EDO, FAD, SO4 |
 |
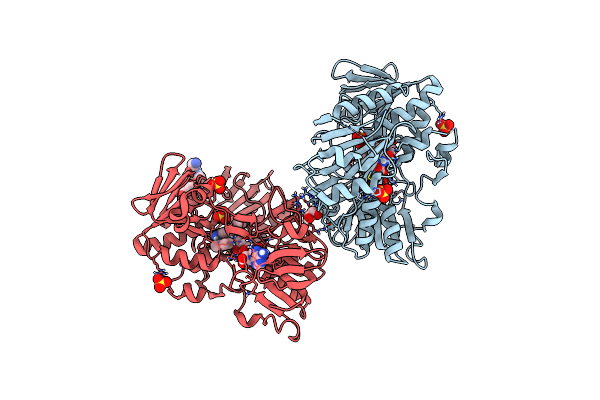
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 7-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QBC, FAD, EDO, SO4 |
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 8-Methoxyquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QF6, EDO, FAD, SO4 |
 |
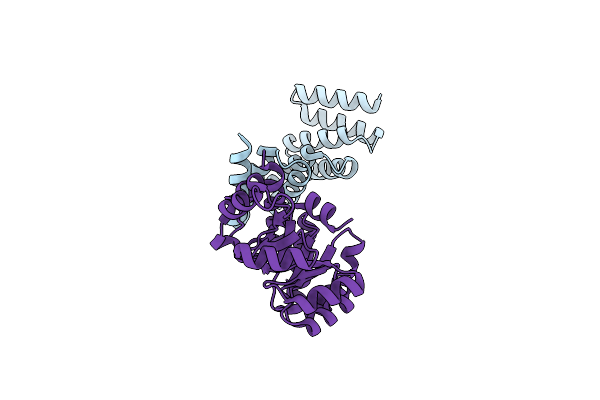
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.98 Å Release Date: 2023-11-01 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2023-11-01 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2023-11-01 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-11-01 Classification: DE NOVO PROTEIN |

