Search Count: 32
 |
Structure Of The Pdz1 Domain From Human Pdzk1 (Nherf3) With The C-Terminal Residues (Kstqf) Of Human Urat1 Transporter (Slc22A12)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2026-01-14 Classification: PROTEIN BINDING |
 |
Structure Of The Pdz2 Domain From Human Pdzk1 (Nherf3) With The C-Terminal Residues (Kstqf) Of Human Urat1 Transporter (Slc22A12)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2026-01-14 Classification: PROTEIN BINDING Ligands: EDO |
 |
Structure Of The Pdz4 Domain From Human Pdzk1 (Nherf3) With The C-Terminal Residues (Kstqf) Of Human Urat1 Transporter (Slc22A12)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2026-01-14 Classification: PROTEIN BINDING |
 |
Structure Of The Pdz1 Domain From Human Nherf1 With The C-Terminal Residues (Kstqf) Of Human Urat1 Transporter (Slc22A12)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2026-01-14 Classification: PROTEIN BINDING Ligands: ACT |
 |
Structure Of The Pdz1 Domain From Human Pdzk1 (Nherf3) With The C-Terminal Residues (Vlkstqf) Of Human Urat1 Transporter (Slc22A12)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2026-01-14 Classification: PROTEIN BINDING |
 |
Structure Of The Pdz4 Domain From Human Pdzk1 (Nherf3) With The C-Terminal Residues (Vlkstqf) Of Human Urat1 Transporter (Slc22A12)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-14 Classification: PROTEIN BINDING Ligands: GOL |
 |
Structure Of The Pdz1 Domain From Human Nherf1 With The C-Terminal Residues (Natrl) Of The Human Sodium-Dependent Phosphate Transporter 2A (Napi-Iia, Slc34A1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2026-01-14 Classification: PROTEIN BINDING Ligands: TOE |
 |
Organism: Yersinia enterocolitica
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2024-05-22 Classification: TRANSFERASE Ligands: UDP, ACT, EDO, CD, MN, NA |
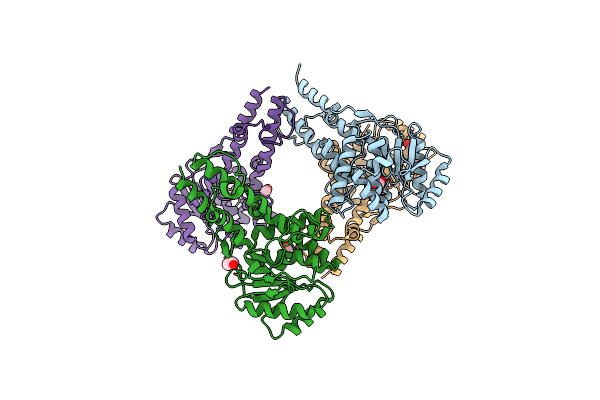 |
Organism: Yersinia enterocolitica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-15 Classification: TRANSFERASE Ligands: EDO, CL |
 |
Cytochrome Bcc-Aa3 Supercomplex (Respiratory Supercomplex Iii2/Iv2) From Corynebacterium Glutamicum (Stigmatellin And Azide Bound)
|
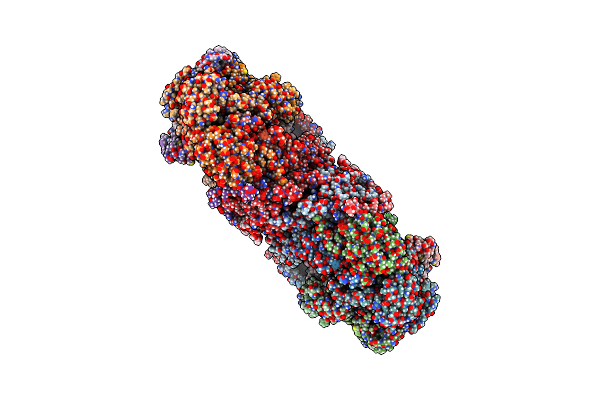 |
Cytochrome Bcc-Aa3 Supercomplex (Respiratory Supercomplex Iii2/Iv2) From Corynebacterium Glutamicum (As Isolated)
|
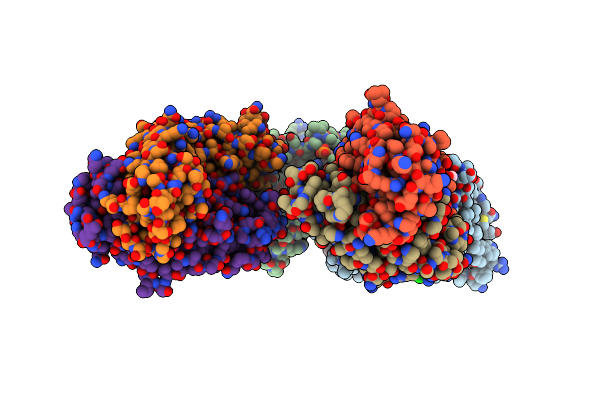 |
Structure Of Dimeric Sodium Proton Antiporter Nhaa, At Ph 8.5, Crystallized With Chimeric Fab Antibodies
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2, Gallus gallus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-02-23 Classification: MEMBRANE PROTEIN Ligands: CDL, 3PH, PO4, PG4, CL |
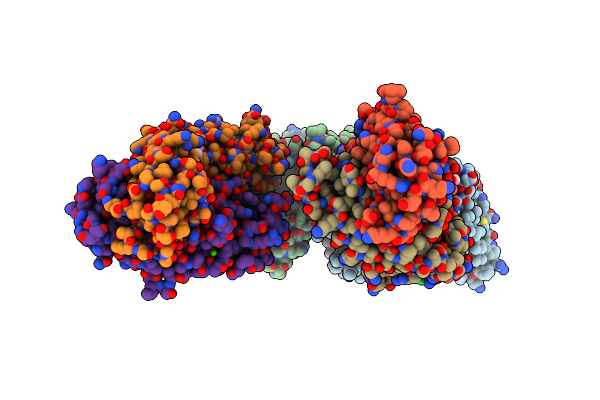 |
Structure Of Dimeric Sodium Proton Antiporter Nhaa, At Ph 6.0, Crystallized With Chimeric Fab Antibodies
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720), Gallus gallus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2022-02-23 Classification: MEMBRANE PROTEIN Ligands: CDL, 3PH, PO4, PG4, 3PE, CL, CA |
 |
Structure Of Dimeric Sodium Proton Antiporter Nhaa K300R Variant, At Ph 8.2, Crystallized With Chimeric Fab Antibodies
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2, Gallus gallus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-02-23 Classification: MEMBRANE PROTEIN Ligands: CDL, 3PH, PO4, PG4, 3PE, CL |
 |
Crystal Structure Of The Udp-Bound Glycosyltransferase Domain From The Ygt Toxin
Organism: Yersinia mollaretii (strain atcc 43969 / dsm 18520 / cip 103324 / cny 7263 / waip 204)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-03-18 Classification: TOXIN Ligands: UDP, MN, K, EDO |
 |
Crystal Structure Of The Ligand-Free Glycosyltransferase Domain From The Ygt Toxin
Organism: Yersinia mollaretii (strain atcc 43969 / dsm 18520 / cip 103324 / cny 7263 / waip 204)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-03-18 Classification: TOXIN |
 |
Crystal Structure Of A Variant (Q133C In Psst) Of Yarrowia Lipolytica Complex I
Organism: Yarrowia lipolytica
Method: X-RAY DIFFRACTION Resolution:3.79 Å Release Date: 2018-12-26 Classification: OXIDOREDUCTASE Ligands: FES, SF4 |
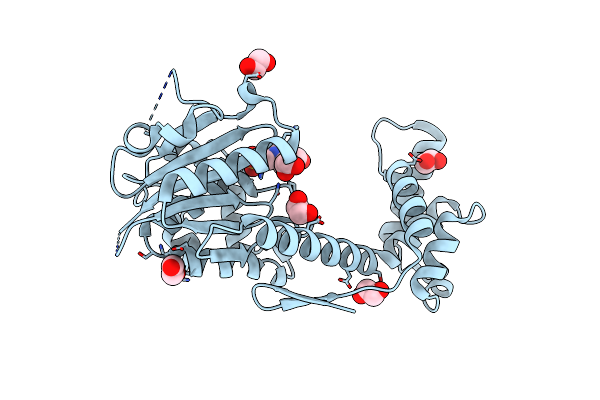 |
Crystal Structure Of Patoxp, A Cysteine Protease-Like Domain Of Photorhabdus Asymbiotica Toxin Patox
Organism: Photorhabdus asymbiotica subsp. asymbiotica atcc 43949
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-05 Classification: TOXIN Ligands: EDO, EPE |
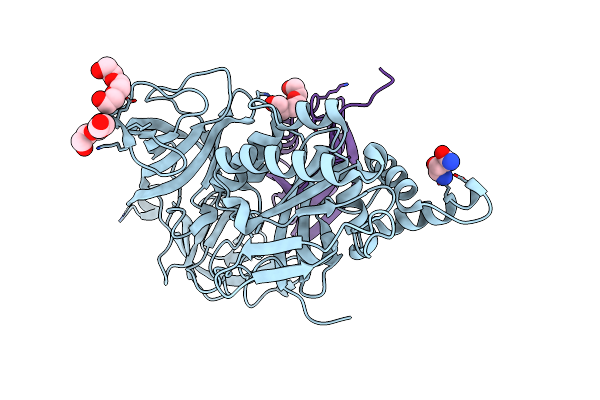 |
Crystal Structure Analysis Of The Yeast Elongation Factor Complex Eef1A:Eef1Ba
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2017-08-23 Classification: TRANSLATION Ligands: GLN, PGE |
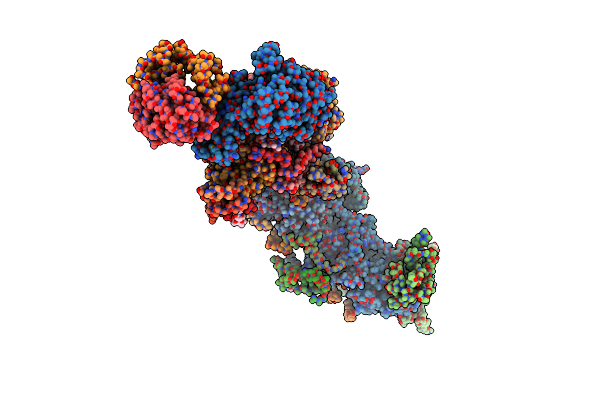 |
Crystal Structure Of Mitochondrial Nadh:Ubiquinone Oxidoreductase From Yarrowia Lipolytica.
Organism: Yarrowia lipolytica
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2015-03-25 Classification: OXIDOREDUCTASE Ligands: FES, SF4 |

