Search Count: 53
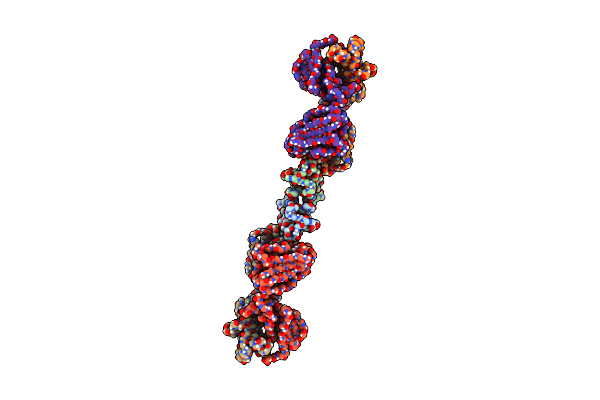 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: LIGASE/IMMUNE SYSTEM |
 |
Human Cx36/Gjd2 Gap Junction Channel With Pore-Lining N-Terminal Helices In Porcine Brain Lipids.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3, Y01 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3, ACD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3, HE2 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: YMZ, MC3 |
 |
Human Cx36/Gjd2 (Ala14-Deleted Mutant) Gap Junction Channel In Porcine Brain Lipids
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Human Cx36/Gjd2 (Ala14 Deletion Mutant) Gap Junction Channel Prepared With Mefloquine, Showing No Bound Mefloquine
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Structure Of The P1B7 Antibody Bound To The Sotorasib-Modified Kras G12C Peptide Presented By The A*03:01 Mhc I Complex
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: IMMUNE SYSTEM Ligands: MOV |
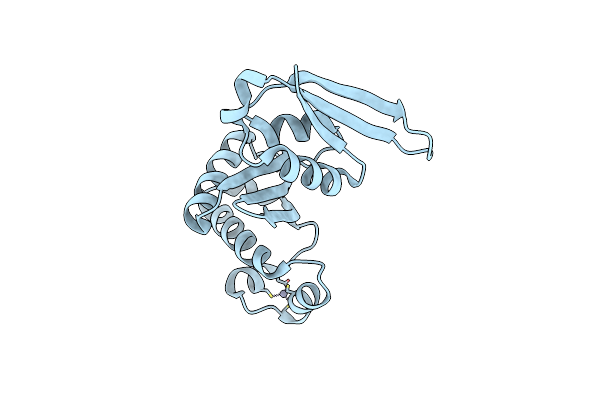 |
Organism: Saccharolobus solfataricus p2
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-10-02 Classification: RNA BINDING PROTEIN Ligands: ZN |
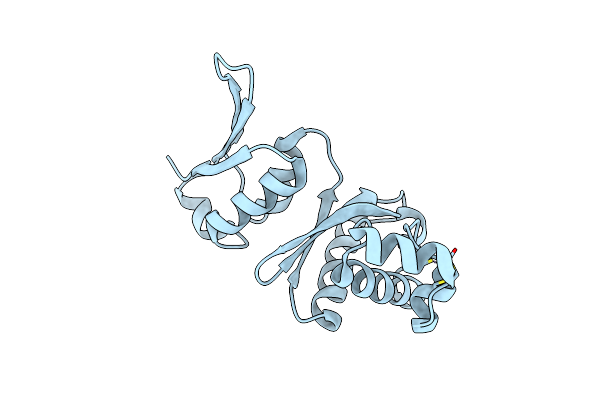 |
Organism: Sulfolobus acidocaldarius dsm 639
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-09-11 Classification: UNKNOWN FUNCTION Ligands: ZN |
 |
Organism: Bos taurus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-09-20 Classification: IMMUNE SYSTEM Ligands: GOL, MES |
 |
Organism: Bos taurus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2023-09-20 Classification: IMMUNE SYSTEM |
 |
Organism: Bos taurus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2023-09-20 Classification: IMMUNE SYSTEM Ligands: PO4 |
 |
Organism: Bos taurus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2023-09-20 Classification: IMMUNE SYSTEM Ligands: GOL, PO4 |
 |
Organism: Bos taurus, Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-09-20 Classification: IMMUNE SYSTEM Ligands: NAG |
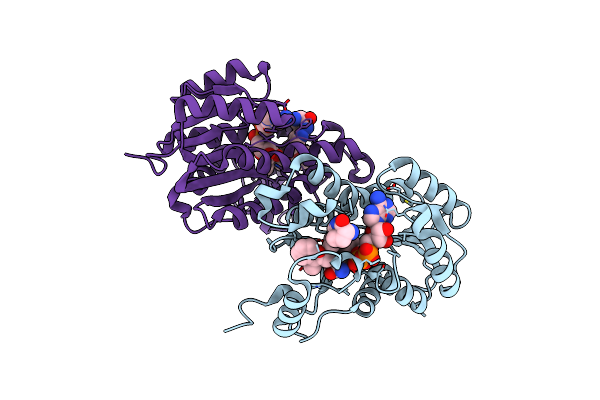 |
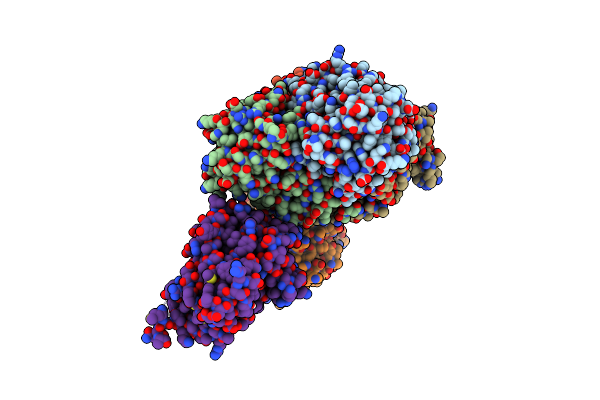
Crystal Structure Of E. Coli Fabi In Complex With Nad And (R,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NAD, NQF |
 |
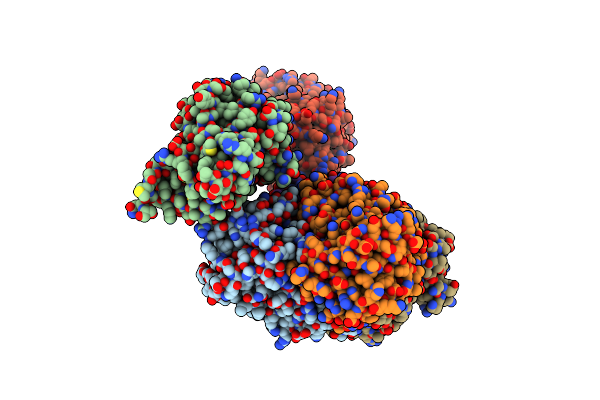
Crystal Structure Of Acinetobacter Baumannii Fabi In Complex With Nad And (R,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NQF, NAD |
 |
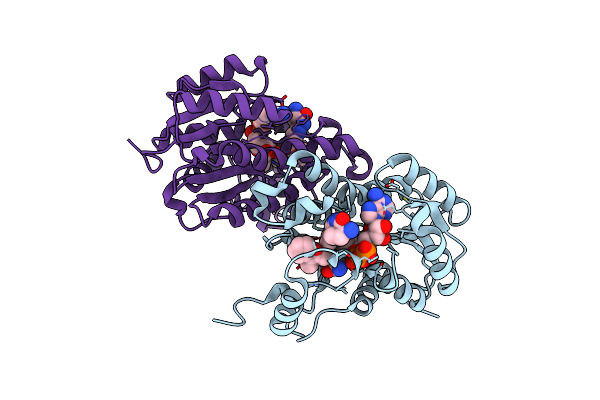
Crystal Structure Of Acinetobacter Baumannii Fabi In Complex With Nad And Fabimycin ((S,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide)
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2023-02-15 Classification: ANTIBIOTIC Ligands: NAD, NUC |
 |
Crystal Structure Of E. Coli Fabi In Complex With Nad And Fabimycin ((S,E)-3-(7-Amino-8-Oxo-6,7,8,9-Tetrahydro-5H-Pyrido[2,3-B]Azepin-3-Yl)-N-Methyl-N-((3-Methylbenzofuran-2-Yl)Methyl)Acrylamide)
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-12-14 Classification: ANTIBIOTIC Ligands: NAD, NUC |

