Search Count: 16
 |
Streptomyces Spheroides Novo (8-Demethylnovbiocic Acid Methyltransferase) With Sah
Organism: Streptomyces niveus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-01-18 Classification: TRANSFERASE Ligands: SAH, GOL, ACT |
 |
Crystal Structure Of The Apo Form Of Thermus Thermophilus Malate Dehydrogenase
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-02-05 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Thermus Thermophilus Malate Dehydrogenase In Complex With Nad
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2014-02-05 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2013-06-12 Classification: HYDROLASE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-01-12 Classification: HYDROLASE |
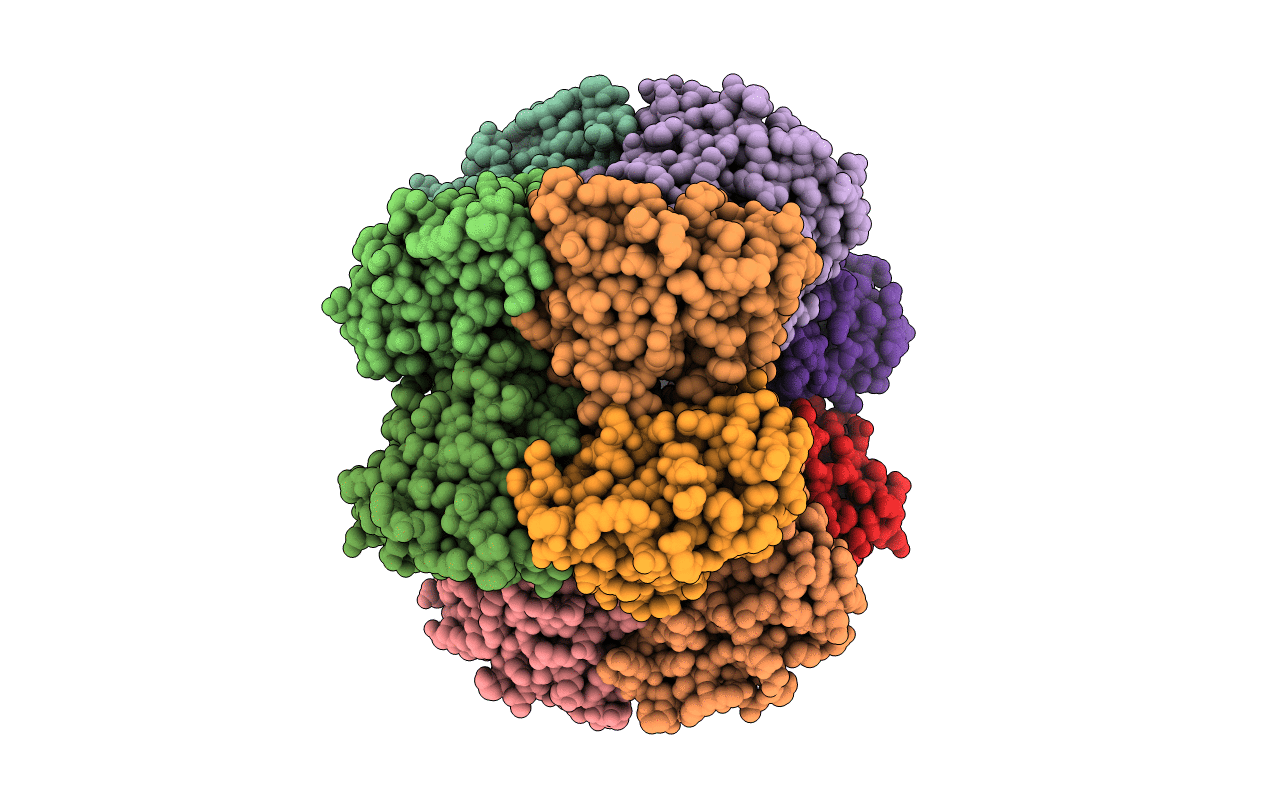 |
Crystal Structure Of Bacillus Subtilis Codw, A Non-Canonical Hslv-Like Peptidase With An Impaired Catalytic Apparatus
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-03-25 Classification: HYDROLASE |
 |
Crystal Structure Of Bacillus Subtilis Codw, A Non-Canonical Hslv-Like Peptidase With An Impaired Catalytic Apparatus
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-03-25 Classification: HYDROLASE |
 |
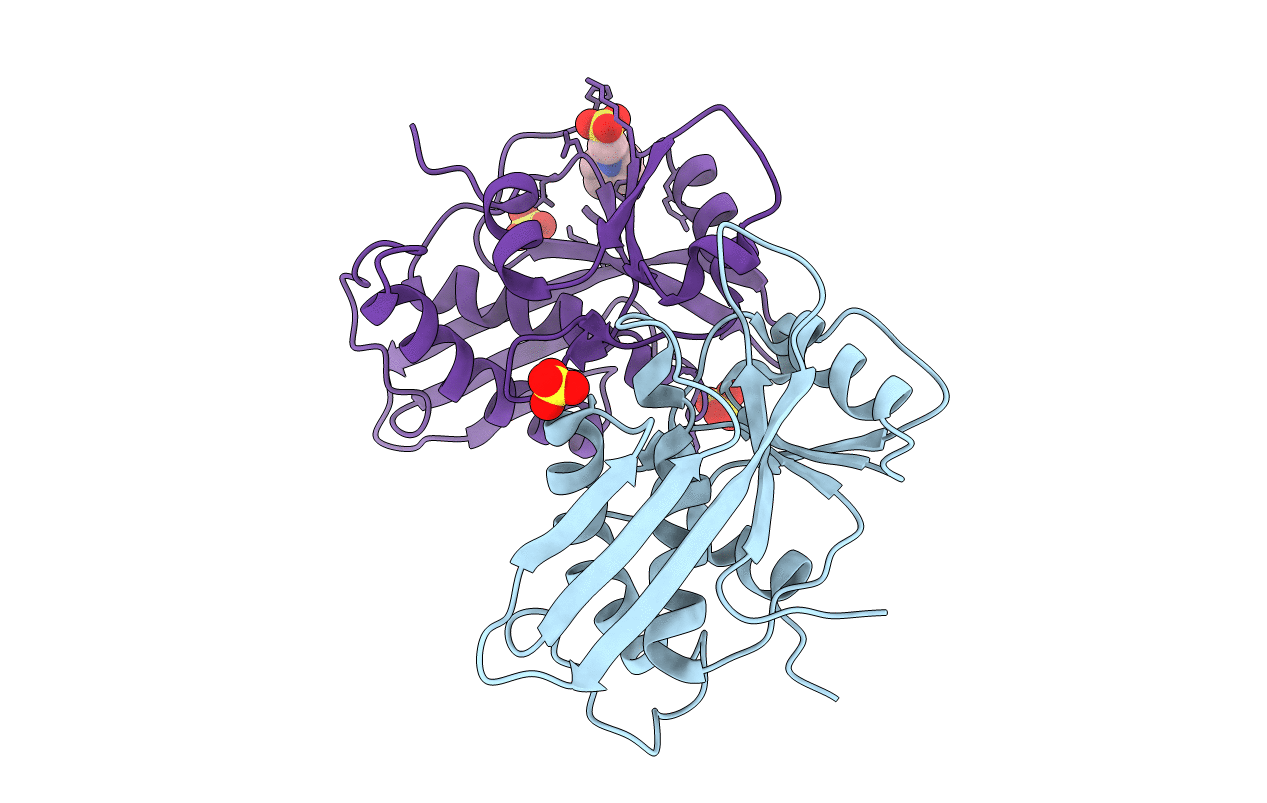
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-03-04 Classification: HYDROLASE |
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: SO4, MES |
 |
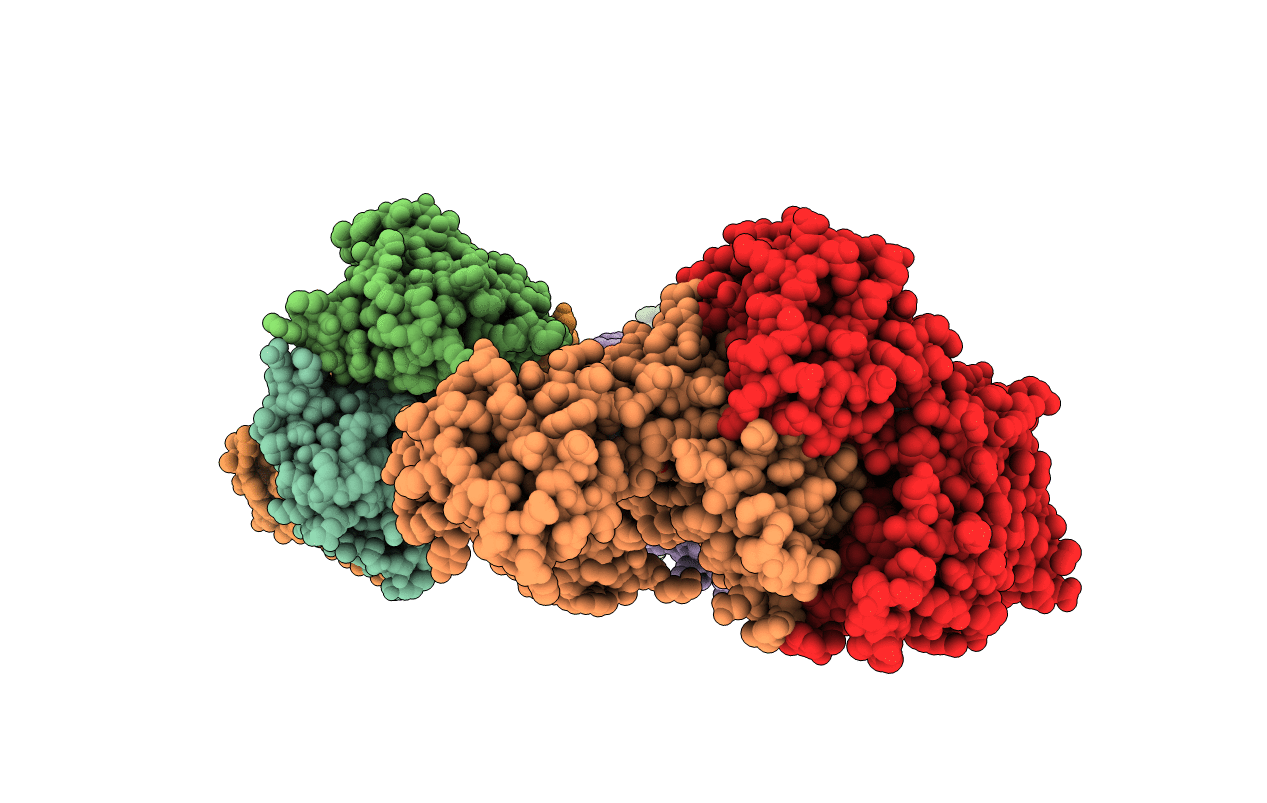
Nucleotide-Dependent Conformational Changes In A Protease-Associated Atpase Hslu
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2001-11-14 Classification: CHAPERONE Ligands: ADP |
 |
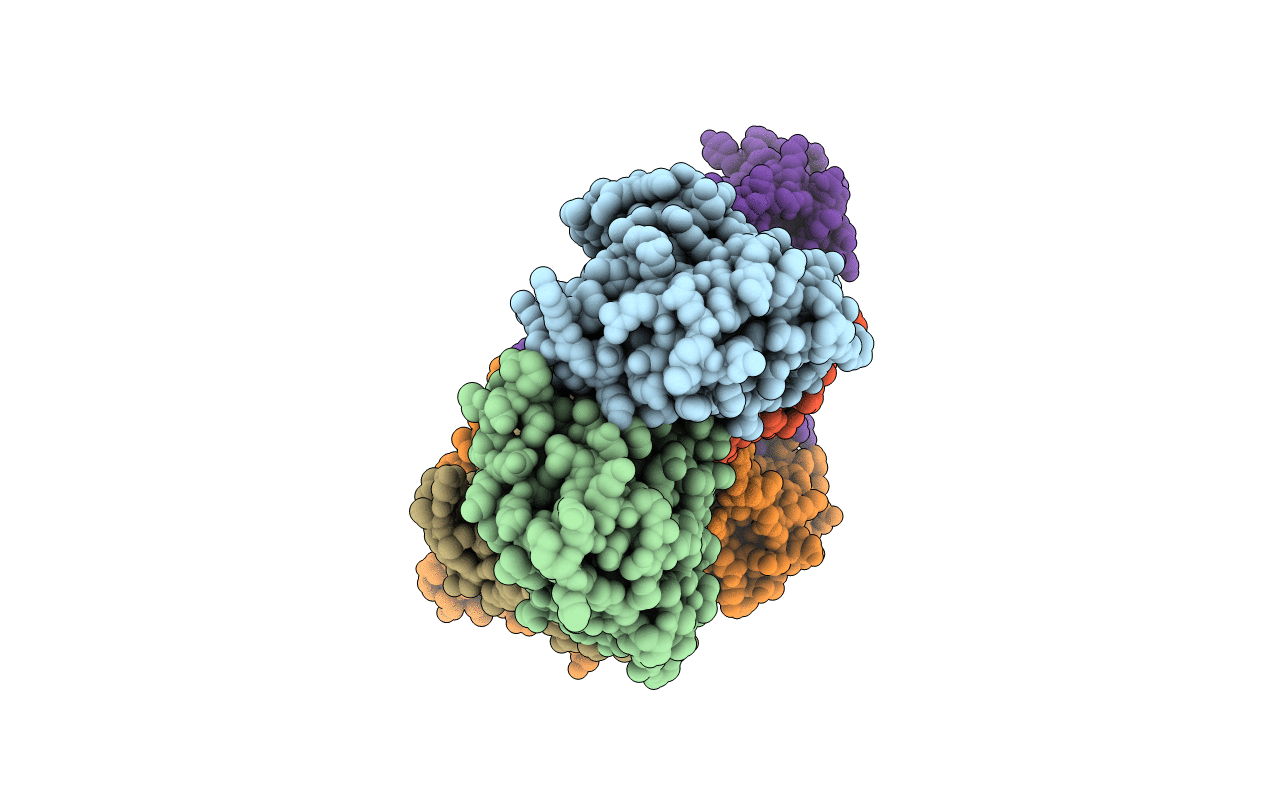
Nucleotide-Dependent Conformational Changes In A Protease-Associated Atpase Hslu
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2001-11-14 Classification: CHAPERONE Ligands: ADP |
 |
Nucleotide-Dependent Conformational Changes In A Protease-Associated Atpase Hslu
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2001-11-14 Classification: CHAPERONE Ligands: ADP |
 |
Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2001-02-21 Classification: CHAPERONE/HYDROLASE Ligands: DAT |
 |
Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:7.00 Å Release Date: 2001-02-21 Classification: CHAPERONE/HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 1997-02-12 Classification: PROTEASE INHIBITOR Ligands: GLC, BGC |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 1997-02-12 Classification: PROTEASE INHIBITOR Ligands: BOG |

