Search Count: 144
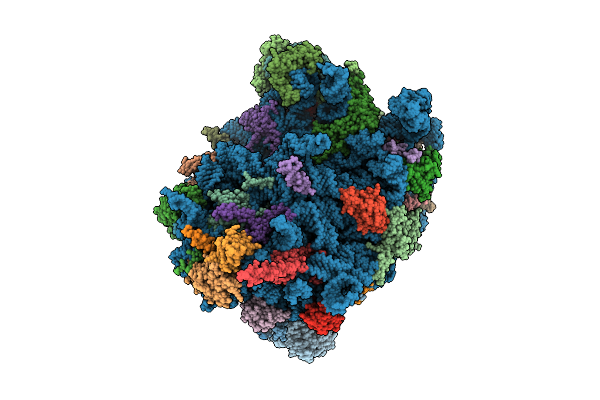 |
Cryo-Em Structure Of Chaetomium Thermophilum Ribosome-Bound Snd3 Translocon
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
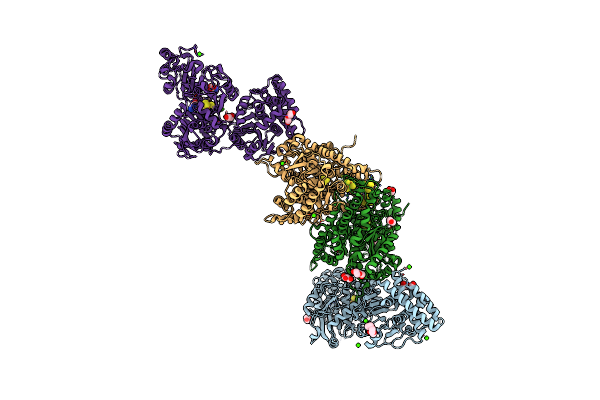 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Acetyl-Coenyzme A From Clostridium Autoethanogenum
Organism: Clostridium autoethanogenum dsm 10061
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: SF4, NI, EDO, PE4, GOL, PEG, CA, CL, XCC, ACO, TRS |
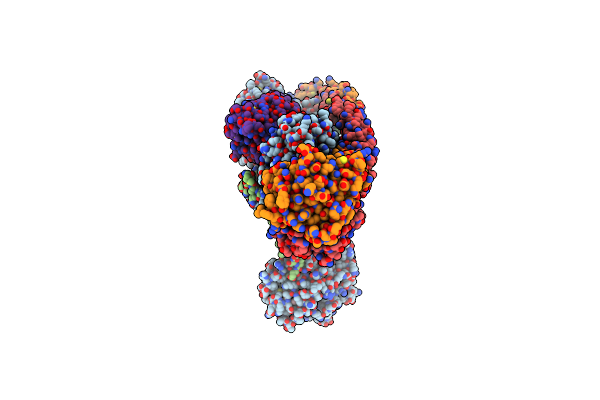 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: SIGNALING PROTEIN Ligands: GTP, MG, MYR |
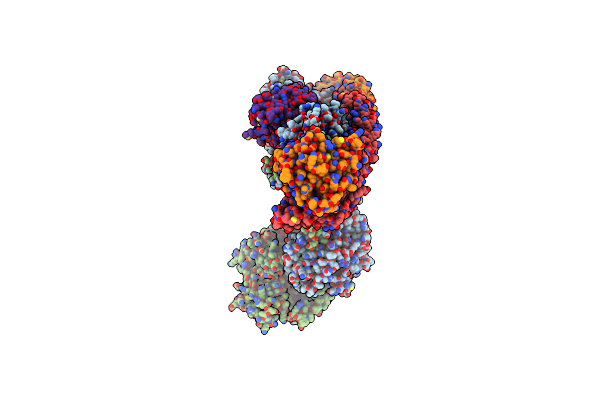 |
Cryo Em Reconstruction Of Pi3Kc3-C1 In Complex With Human Rab1A(Q70L), Vps34 Kinase Domain In The Inactive Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: SIGNALING PROTEIN Ligands: GTP, MG, MYR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: SIGNALING PROTEIN Ligands: GTP, MG |
 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3A)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, B12, NI, RQM |
 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3B)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, RQM, B12 |
 |
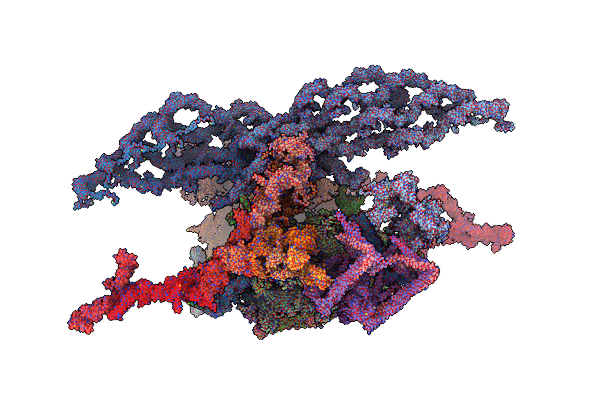
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3Cb)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, B12, RQM |
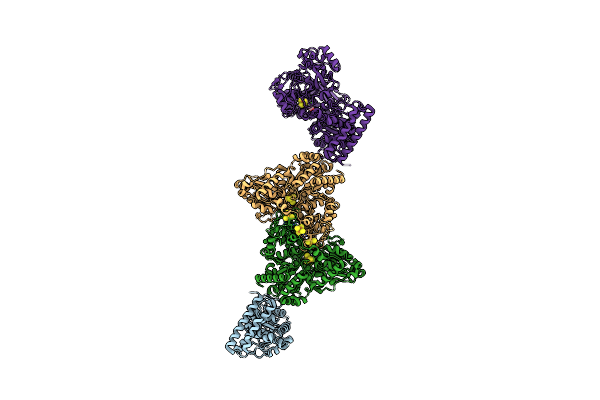 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) From Clostridium Autoethanogenum (Composite Structure, Closed And Co-Bound State)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, RQM, NI, CMO |
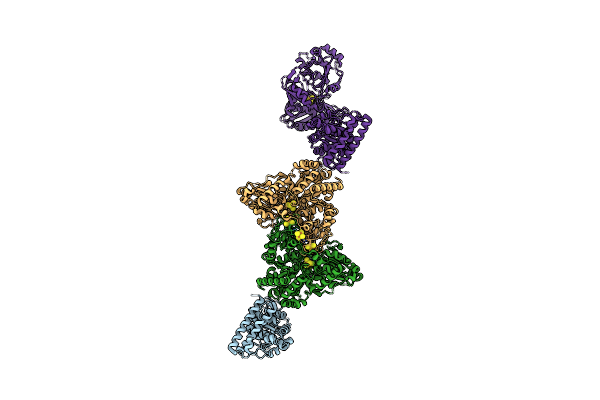 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) From Clostridium Autoethanogenum (Composite Structure, Semi-Extended State)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, RQM |
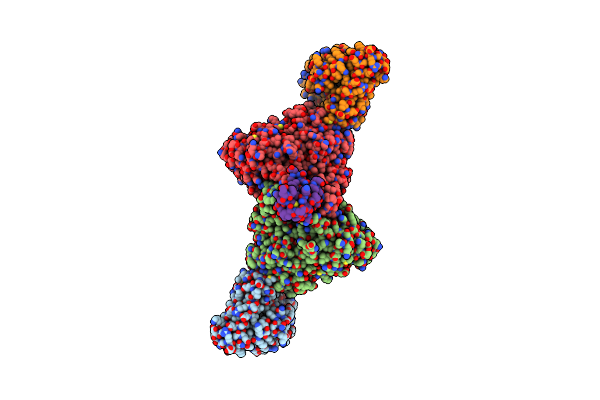 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Ferredoxin (Clostridium Autoethanogenum)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, RQM |
 |
Organism: Dictyostelium discoideum
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: NUCLEAR PROTEIN |
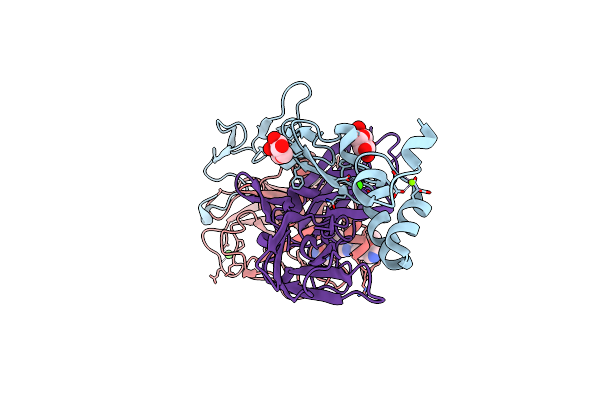 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-10-16 Classification: BLOOD CLOTTING Ligands: CA, MG, GLC, FUC, CL, WSV |
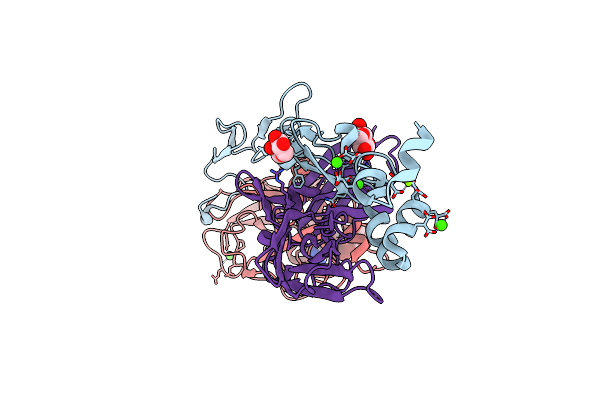 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-10-09 Classification: BLOOD CLOTTING Ligands: CA, MG, GLC, FUC, WIZ, CL |
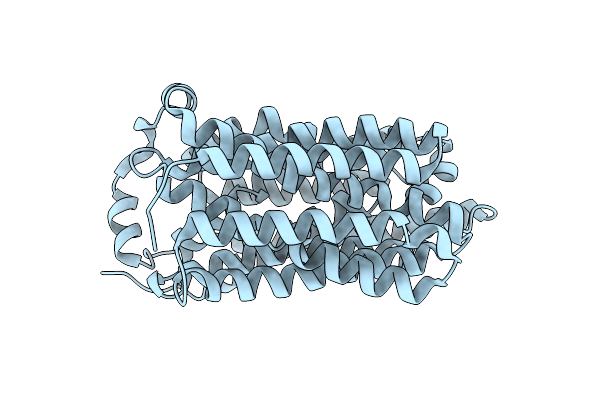 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN |
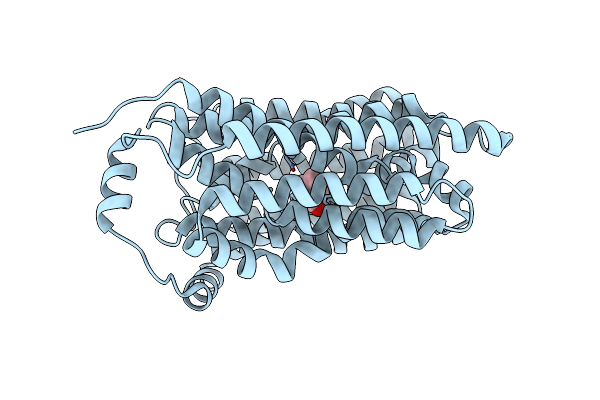 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CHT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CHT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: ETA |

