Search Count: 17
 |
Crystal Structure Of Methylmalonyl-Coa Epimerase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-12 Classification: ISOMERASE Ligands: SO4 |
 |
The Structure Of Archaeal Inorganic Pyrophosphatase In Complex With Pyrophosphate
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-02-08 Classification: HYDROLASE Ligands: CA, EDO, MRD, POP, NA |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2012-12-12 Classification: HYDROLASE |
 |
The Structure Of Manganese Bound Thermococcus Thioreducens Inorganic Pyrophosphatase
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2012-03-28 Classification: HYDROLASE Ligands: MN |
 |
The Structure Of Calcium Bound Thermococcus Thioreducens Inorganic Pyrophosphatase At 298K
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-03-28 Classification: HYDROLASE Ligands: CA, MRD, MPD |
 |
The Structure Of Thermococcus Thioreducens' Inorganic Pyrophosphatase Bound To Sulfate
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2012-03-28 Classification: HYDROLASE Ligands: SO4, NH4 |
 |
The Neutron Crystallographic Structure Of Inorganic Pyrophosphatase From Thermococcus Thioreducens
Organism: Thermococcus thioreducens
Method: NEUTRON DIFFRACTION Resolution:2.50 Å Release Date: 2012-02-08 Classification: HYDROLASE Ligands: CA, DOD |
 |
Magnesium Activated Inorganic Pyrophosphatase From Thermococcus Thioreducens Bound To Hydrolyzed Product At 0.99 Angstrom Resolution
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2012-01-04 Classification: HYDROLASE Ligands: PO4, MG, EPE, CL |
 |
The Structure Of Archaeal Inorganic Pyrophosphatase In Complex With Substrate
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2012-01-04 Classification: HYDROLASE Ligands: CA, POP, BR, NA |
 |
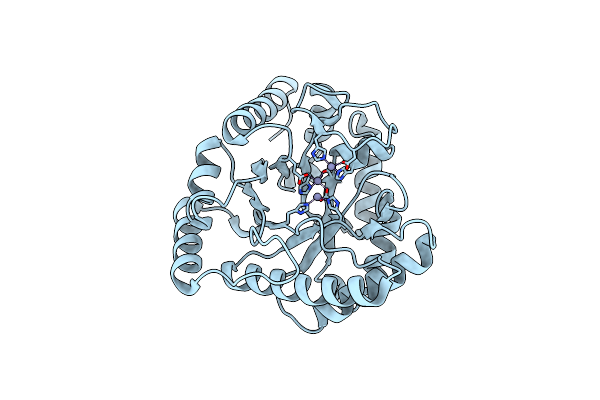
The Structure Of Inorganic Pyrophosphatase From Thermococcus Thioreducens In Complex With Magnesium And Sulfate
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2012-01-04 Classification: HYDROLASE Ligands: MG, SO4 |
 |
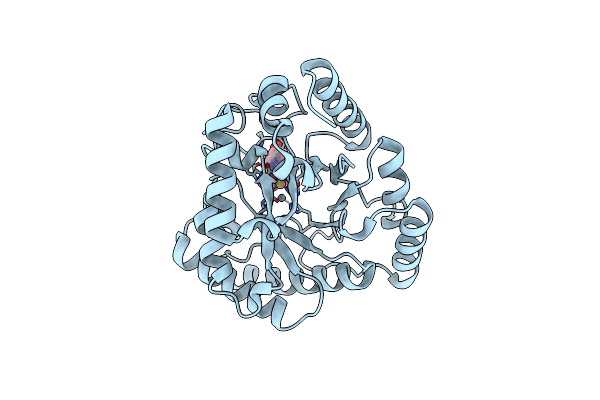
Crystal Structure Of Thermotoga Maritima Endonuclease Iv In The Presence Of Zinc
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-09-08 Classification: HYDROLASE Ligands: ZN |
 |
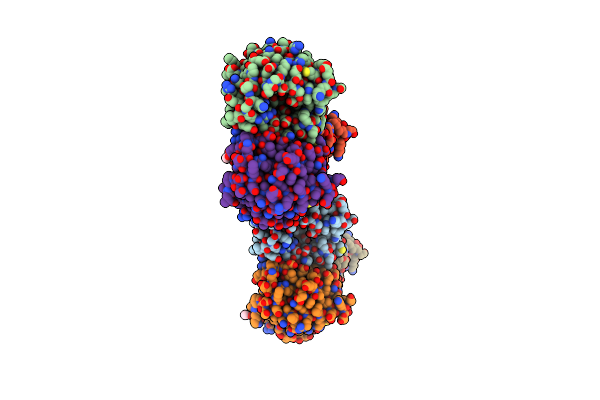
Crystal Structure Of Thermotoga Maritima Endonuclease Iv In The Presence Of Cadmium And Zinc
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2010-09-08 Classification: HYDROLASE Ligands: ZN, CD, BCN |
 |
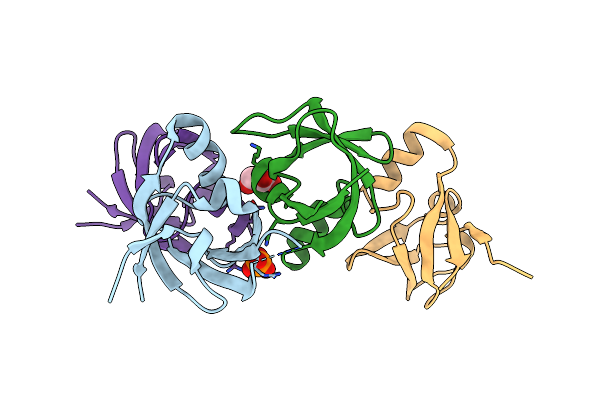
X-Ray Crystallographic Structure Of Inorganic Pyrophosphatase At 298K From Archaeon Thermococcus Thioreducens
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-08-18 Classification: HYDROLASE Ligands: MRD, CA, MPD, ACE |
 |
Crystal Structure Of Iron Uptake Regulatory Protein (Feoa) Solved By Sulfur Sad In A Monoclinic Space Group
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-12-16 Classification: TRANSCRIPTION REGULATOR, METAL BINDING PROTEIN Ligands: PO4, GOL |
 |
Crystal Structure Of Ubiquitin-Conjugating Enzyme E2-25Kda (Huntington Interacting Protein 2) M172A Mutant Crystallized At Ph 8.5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2008-11-25 Classification: LIGASE Ligands: PG4, BME, TRS, CA |
 |
Crystal Structure Of Ubiquitin-Conjugating Enzyme E2-25Kda (Huntington Interacting Protein 2) M172A Mutant
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2008-08-26 Classification: LIGASE Ligands: CA |
 |
Crystal Structure Of Piwi/Argonaute/Zwille(Paz) Domain From Thermococcus Thioreducens
Organism: Thermococcus thioreducens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2008-07-29 Classification: RNA BINDING PROTEIN |

