Search Count: 43
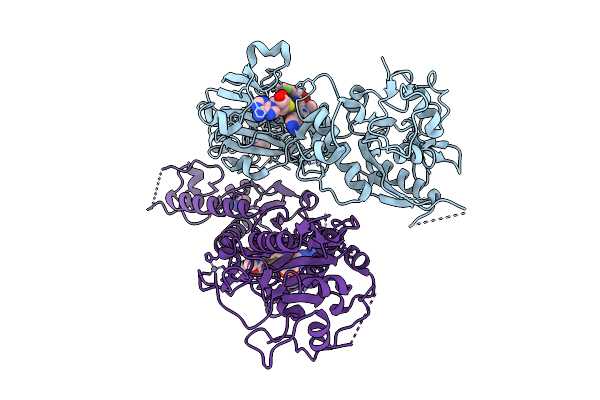 |
Organism: Sars-cov-2 pseudovirus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: VIRAL PROTEIN Ligands: ZN, SAH, A1JLH, CL, IMD |
 |
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: VIRAL PROTEIN Ligands: ZN, SAH, A1JDF, CL, EDO, IMD, IPA, SO4 |
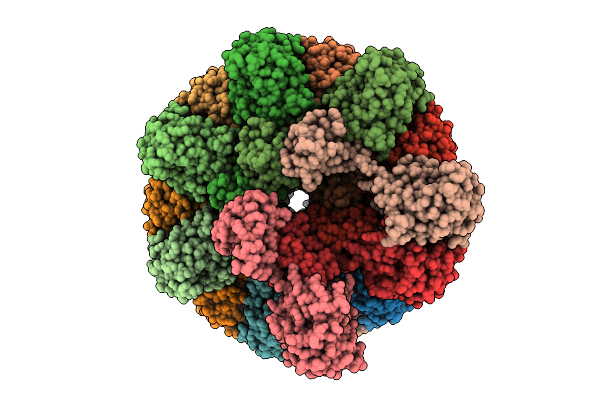 |
Cryo-Em Structure Of Human Tankyrase 2 Sam-Parp Filament Bound To Compound, Tdi-2804 (Consensus Map).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: ZN, A1AE4 |
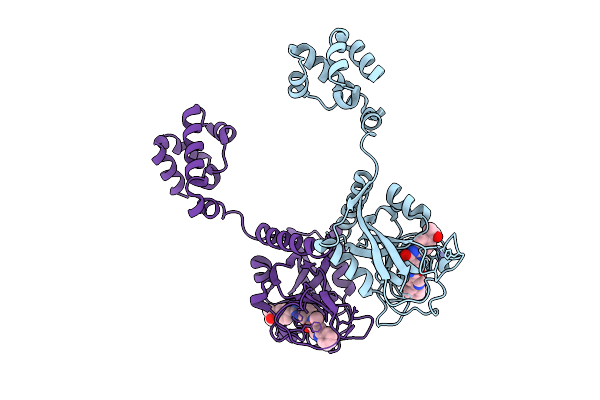 |
Cryo-Em Structure Of Human Tankyrase 2 Sam-Parp Filament Bound To Compound, Tdi-2804 (Focused Refinement Map).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: ZN, A1AE4 |
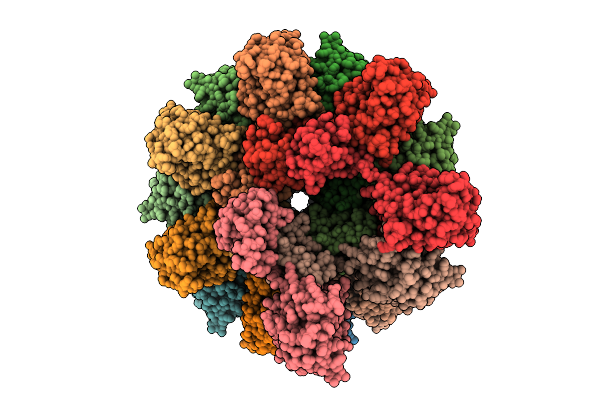 |
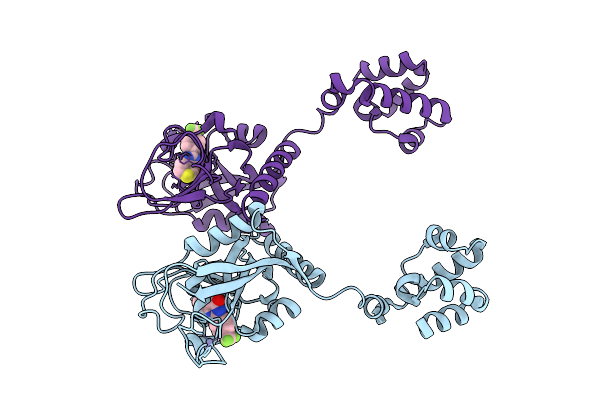
Cryo-Em Structure Of Human Tankyrase 2 Sam-Parp Filament Bound To Compound, Xav (Consensus Map).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: XAV, ZN |
 |
Cryo-Em Structure Of Human Tankyrase 2 Sam-Parp Filament Bound To Compound, Xav (Focused Refinement Map).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: XAV, ZN |
 |
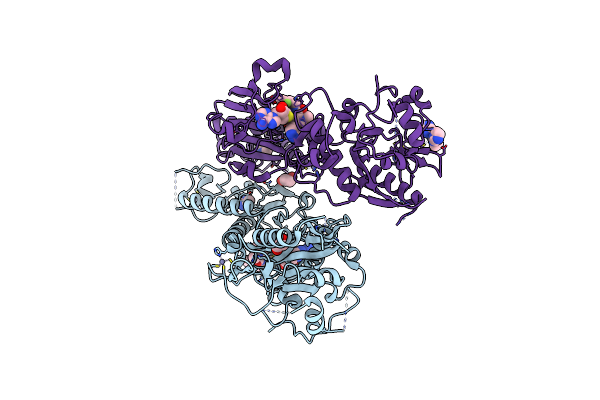
Cryo-Em Structure Of Human Tankyrase 2 Sam-Parp Filament - Apo State (Focused Refinement Map).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: ZN |
 |
Cryo-Em Structure Of Human Tankyrase 2 Sam-Parp Filament -Apo State (Consensus Map).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: ZN |
 |
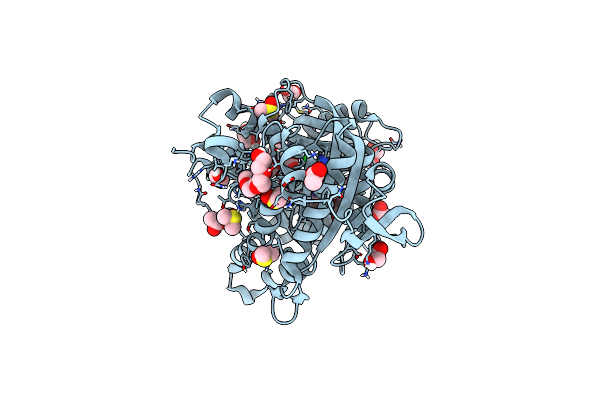
Organism: Sars-cov-2 pseudovirus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-10-23 Classification: VIRAL PROTEIN Ligands: ZN, SAH, EDO, IMD, YDT, CL |
 |
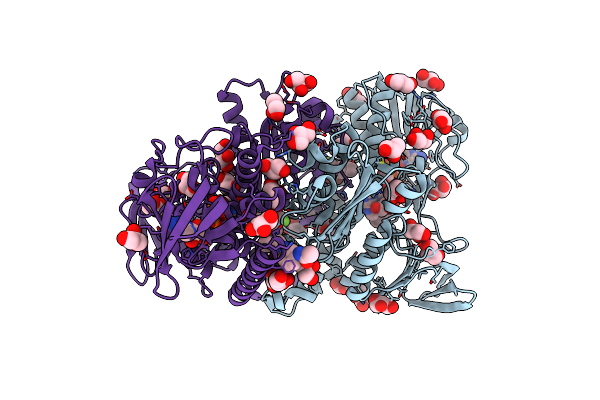
Crystal Structure Of Human Soluble Adenylyl Cyclase (Sac) In Complex With Inhibitor Tdi-09066
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-03 Classification: SIGNALING PROTEIN Ligands: V9E, DMS, ACT, EDO, PG4, PGE, CL |
 |
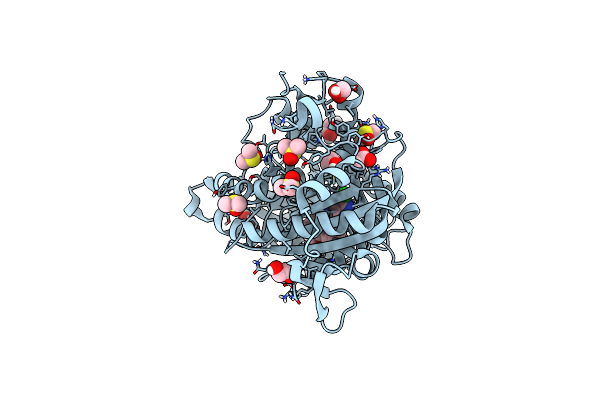
Co-Crystal Structure Of Optimized Analog Tdi-13537 Provided New Insights Into The Potency Determinants Of The Sulfonamide Inhibitor Series
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-01-03 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, U4I, GOL |
 |
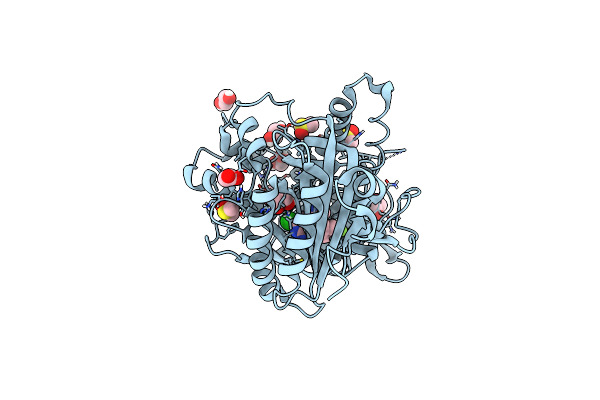
Crystal Structure Of Human Soluble Adenylyl Cyclase (Sac) In Complex With Inhibitor Tdi-10512
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-04-26 Classification: SIGNALING PROTEIN Ligands: V6U, DMS, EDO, ACT |
 |
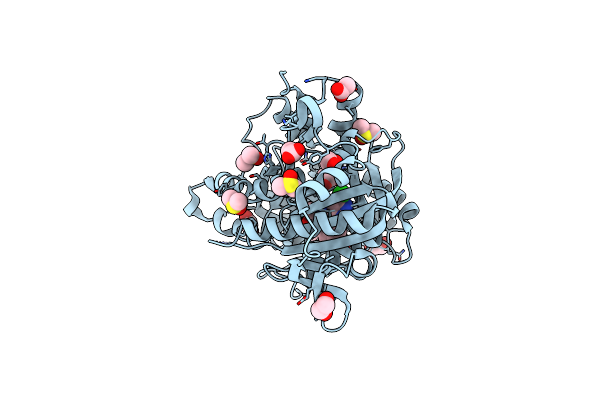
Crystal Structure Of Human Soluble Adenylyl Cyclase Catalytic Domain In Complex With The Inhibitor Tdi-10228
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-04-26 Classification: SIGNALING PROTEIN Ligands: VE1, DMS, ACT, EDO |
 |
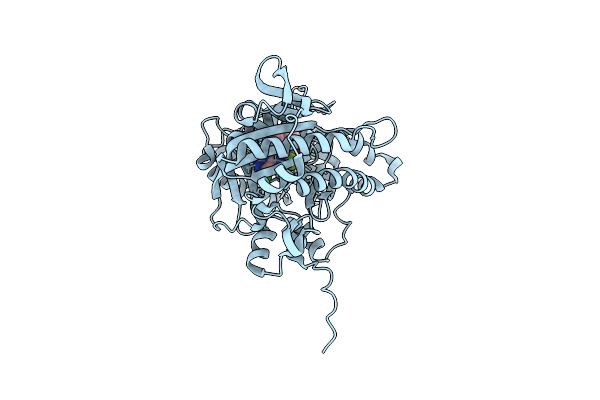
Complex Of Human Soluble Adenylyl Cyclase 10 Catalytic Core With Inhibitor Tdi-10962
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-04-26 Classification: SIGNALING PROTEIN Ligands: VBB, DMS, EDO, ACT |
 |
Crystal Structure Of Human Soluble Adenylyl Cyclase In Complex With The Inhibitor Tdi-011861
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-03-29 Classification: SIGNALING PROTEIN Ligands: PJU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-10-06 Classification: SIGNALING PROTEIN Ligands: 1S2, ACT, DMS, EDO |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-01-27 Classification: DNA BINDING PROTEIN Ligands: NYE, PO4, GOL |
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-01-27 Classification: DNA BINDING PROTEIN Ligands: PO4, O0E |
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2021-01-27 Classification: DNA BINDING PROTEIN Ligands: NZW, CA |
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-01-27 Classification: DNA BINDING PROTEIN Ligands: O08, CA, GOL |

