Search Count: 35
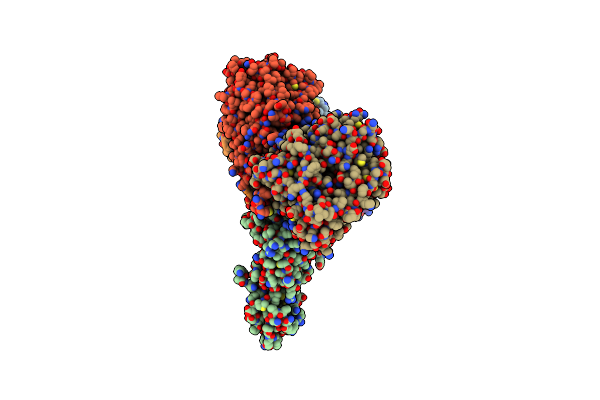 |
Cryo-Em Structure Of Sodium Pumping Rnf Complex From Acetobacterium Woodii Bound To Nadh
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: FES, SF4, FMN, NAD, RBF |
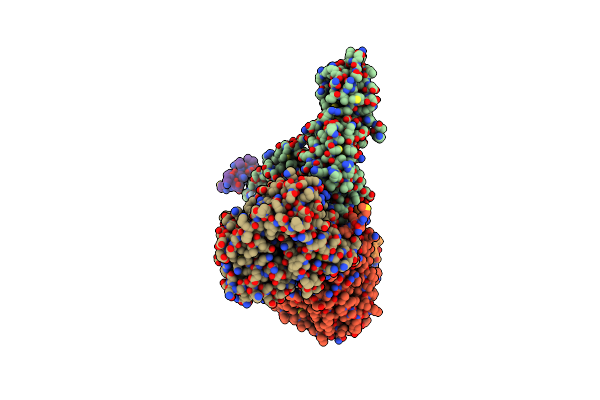 |
Cryo-Em Structure Of Sodium Pumping Rnf Complex From Acetobacterium Woodii Reduced With Low Potential Ferredoxin
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
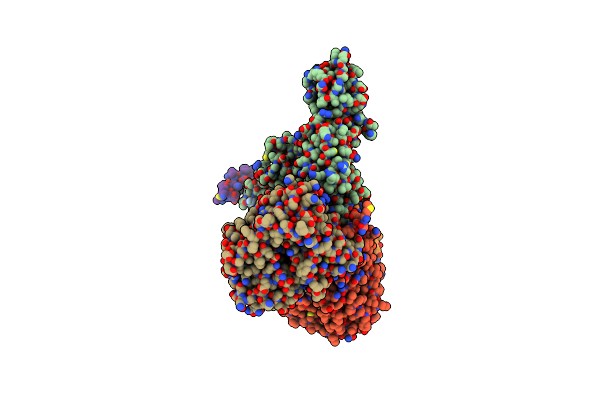 |
Cryo-Em Structure Of Sodium Pumping Rnf Complex From Acetobacterium Woodii Reduced With Low Potential Ferredoxin (Consensus Map)
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
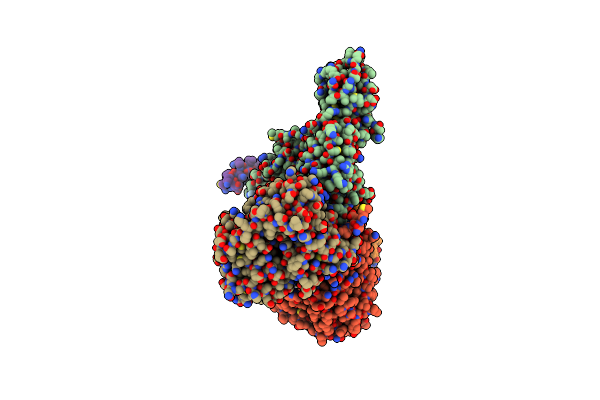 |
Cryo-Em Structure Of Sodium Pumping Rnf Complex From Acetobacterium Woodii In Apo State
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-13 Classification: PROTEIN BINDING Ligands: GOL, LI |
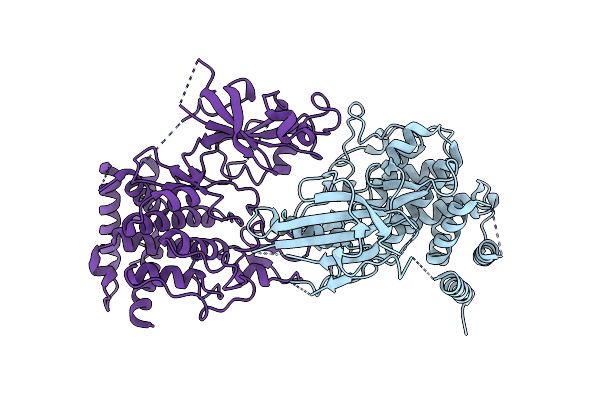 |
Structure Of Dimeric Phosphorylated Pediculus Humanus (Ph) Pink1 With Kinked Alpha-C Helix In Chain B
Organism: Pediculus humanus corporis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSFERASE |
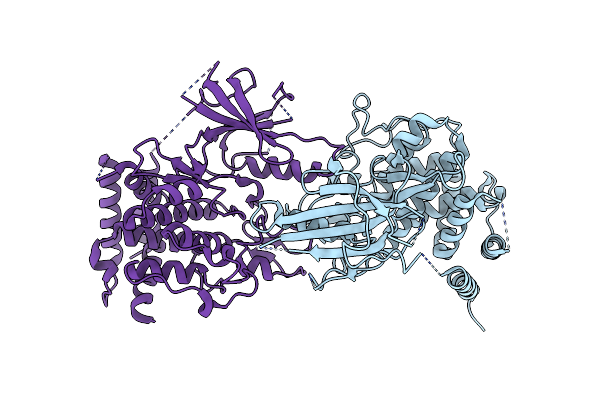 |
Structure Of Dimeric Phosphorylated Pediculus Humanus (Ph) Pink1 With Extended Alpha-C Helix In Chain B
Organism: Pediculus humanus corporis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSFERASE |
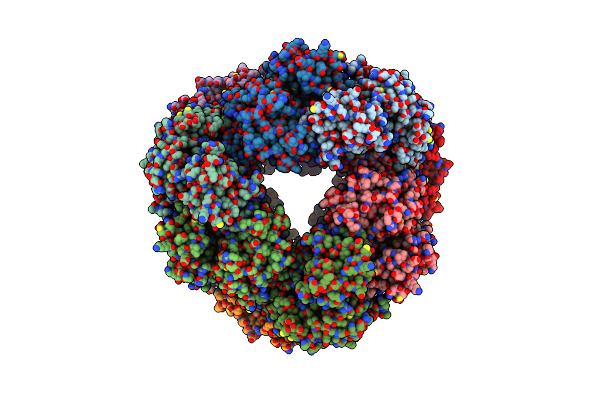 |
Structure Of Dodecameric Unphosphorylated Pediculus Humanus (Ph) Pink1 D357A Mutant
Organism: Pediculus humanus corporis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSFERASE |
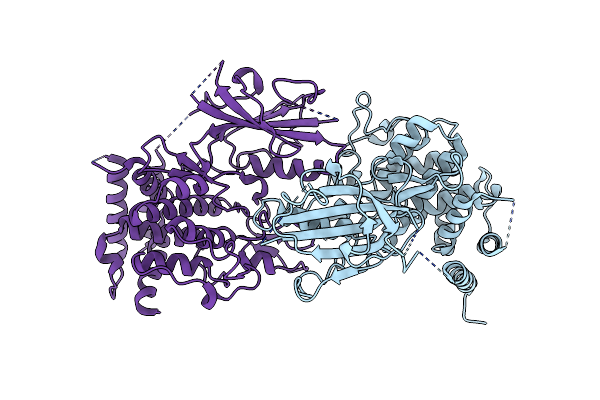 |
Structure Of Dimeric Unphosphorylated Pediculus Humanus (Ph) Pink1 D357A Mutant
Organism: Pediculus humanus corporis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSFERASE |
 |
Organism: Pediculus humanus corporis
Method: X-RAY DIFFRACTION Resolution:3.53 Å Release Date: 2021-12-22 Classification: TRANSFERASE |
 |
Organism: Wolbachia pipientis wmel
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-07-01 Classification: HYDROLASE Ligands: ACT |
 |
Crystal Structure Of An Otu Deubiquitinase From Wolbachia Pipientis Wmel Bound To Ubiquitin
Organism: Homo sapiens, Wolbachia pipientis wmel
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2020-07-01 Classification: HYDROLASE Ligands: FLC |
 |
Crystal Structure Of An Otu Deubiquitinase From Escherichia Albertii Bound To Ubiquitin
Organism: Escherichia albertii (strain tw07627), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-01 Classification: HYDROLASE Ligands: FMT |
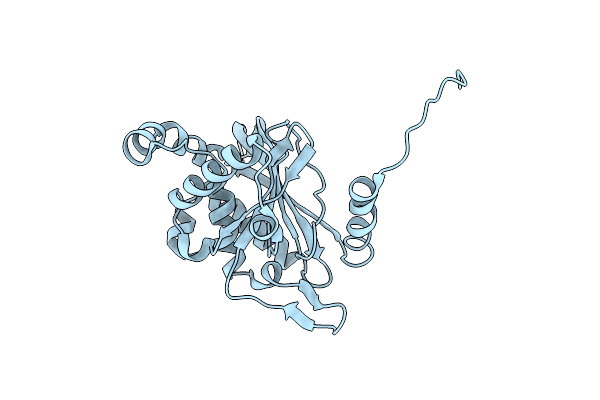 |
Cryo-Electron Microscopic Structure Of The Dihydrolipoamide Succinyltransferase (E2) Component Of The Human Alpha-Ketoglutarate (2-Oxoglutarate) Dehydrogenase Complex [Residues 218-453]
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-01-22 Classification: TRANSFERASE |
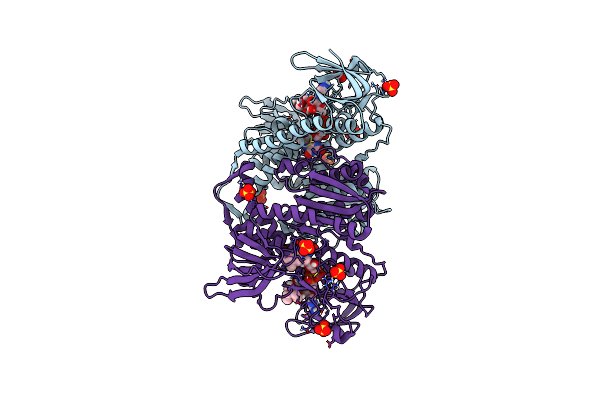 |
Crystal Structure Of The Disease-Causing G194C Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, BTB |
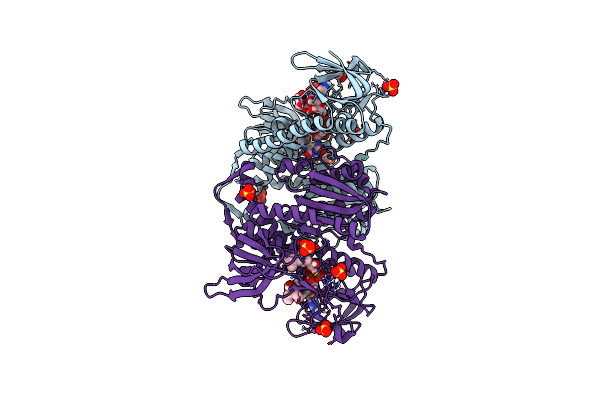 |
Crystal Structure Of The Human Dihydrolipoamide Dehydrogenase At 1.75 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, BTB, SO4 |
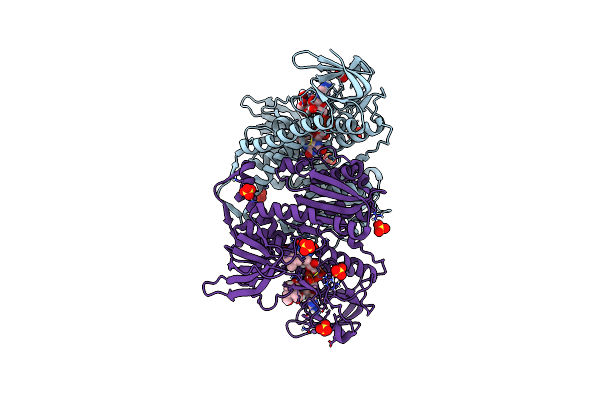 |
Crystal Structure Of The Disease-Causing R460G Mutant Of The Human Dihydrolipoamide Dehydrogenase At 1.44 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, BTB |
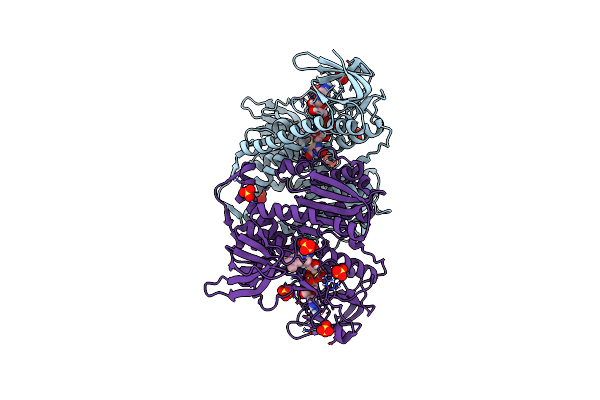 |
Crystal Structure Of The Disease-Causing R447G Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |
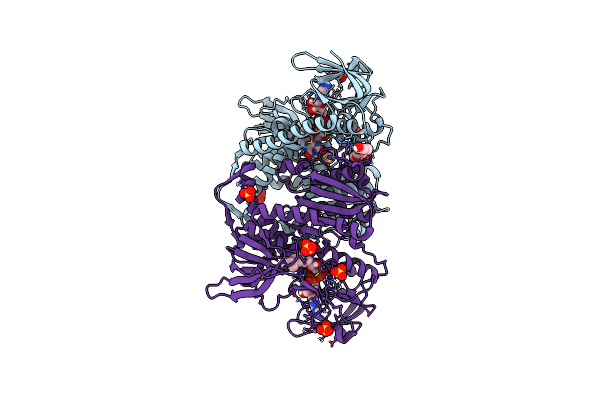 |
Crystal Structure Of The Disease-Causing I445M Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, 1PE |

