Search Count: 17
 |
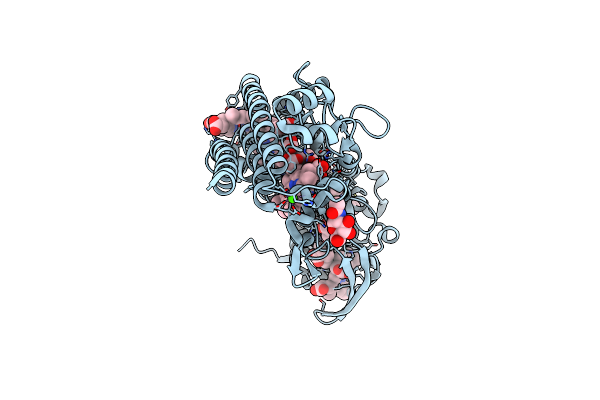
Structure Of Igni18, A Novel Metallo Hydrolase From The Hyperthermophilic Archaeon Ignicoccus Hospitalis Kin4/I
Organism: Ignicoccus hospitalis
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2019-10-09 Classification: HYDROLASE Ligands: ZN, PO4, K |
 |
Organism: Ignicoccus hospitalis kin4/i
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-12-23 Classification: HEME BINDING PROTEIN Ligands: HEC, CA, NAG |
 |
Organism: Homo sapiens
Method: SOLID-STATE NMR Release Date: 2015-02-18 Classification: SIGNALING PROTEIN |
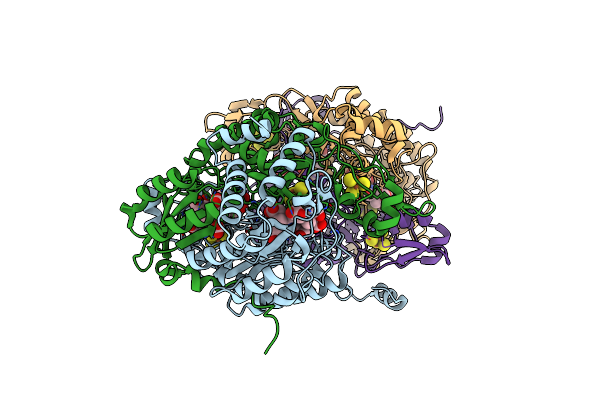 |
Structure Of The Dissimilatory Sulfite Reductase From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2010-05-12 Classification: OXIDOREDUCTASE Ligands: SRM, SF4, GOL |
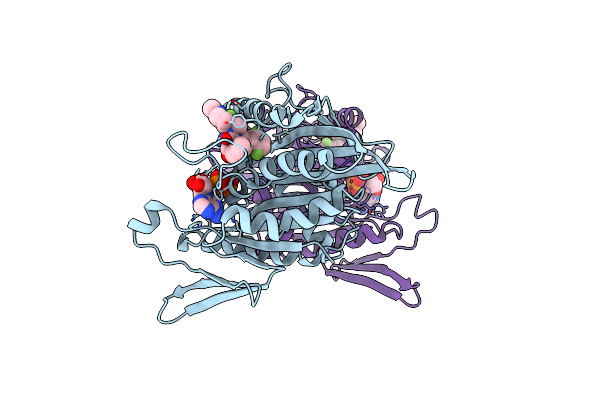 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2008-07-01 Classification: MOTOR PROTEIN Ligands: MG, ADP, K30 |
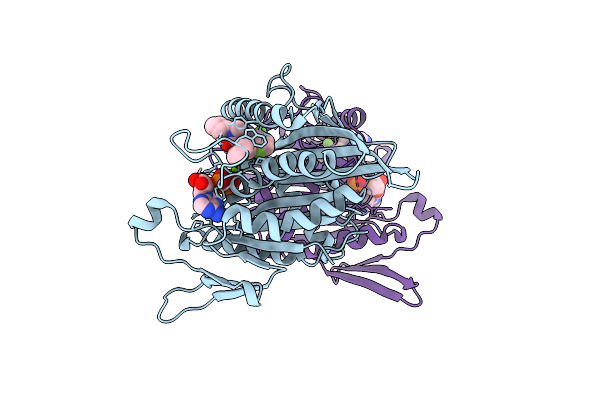 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-02-07 Classification: CELL CYCLE Ligands: MG, ADP, N2T |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-02-07 Classification: CELL CYCLE Ligands: MG, ADP, N4T |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-02-07 Classification: CELL CYCLE Ligands: MG, ADP, N5T |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-04-12 Classification: CELL CYCLE Ligands: MG, ADP, L47 |
 |
Crystal Structure Of The Motor Protein Ksp In Complex With Adp And Monastrol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-01-13 Classification: CELL CYCLE Ligands: MG, ADP, NAT |
 |
Co-Crystal Structure Of Human Farnesyltransferase With Farnesyldiphosphate And Inhibitor Compound 33A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-07-08 Classification: TRANSFERASE Ligands: ZN, FPP, BNE |
 |
Co-Crystal Structure Of Human Farnesyltransferase With Farnesyldiphosphate And Inhibitor Compound 66
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-06-19 Classification: TRANSFERASE Ligands: ZN, FPP, U66 |
 |
Co-Crystal Structure Of Human Farnesyltransferase With Farnesyldiphosphate And Inhibitor Compound 49
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-06-19 Classification: TRANSFERASE Ligands: ACY, ZN, FPP, U49 |
 |
Structure Of Adenylylsulfate Reductase From The Hyperthermophilic Archaeoglobus Fulgidus At 1.6 Resolution
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2002-03-27 Classification: OXIDOREDUCTASE Ligands: SO3, FAD, SF4 |
 |
Structure Of Adenylylsulfate Reductase From The Hyperthermophilic Archaeoglobus Fulgidus At 1.6 Resolution
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2002-02-27 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, SF4 |
 |
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1999-03-23 Classification: CHAPERONIN |
 |
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 1999-03-23 Classification: CHAPERONIN Ligands: MG, ADP, AF3 |

