Search Count: 44
 |
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-06-05 Classification: ANTIMICROBIAL PROTEIN,HYDROLASE Ligands: GFX |
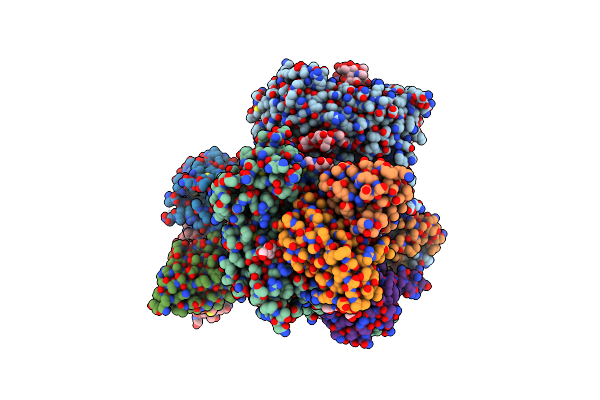 |
Cryo-Em Structure Of Hiv-1 Env Trimer Bg505 Sosip.664 In Complex With Cd4Bs Antibody Ab1573
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
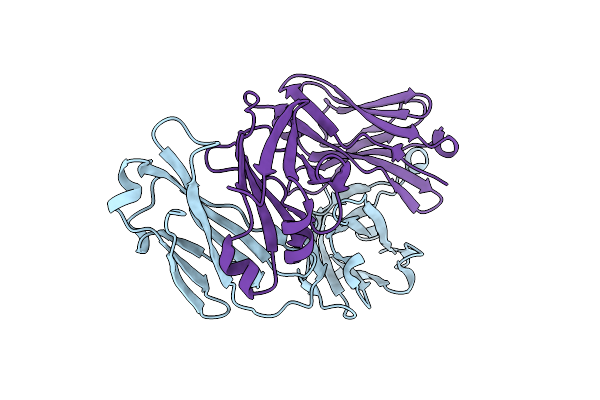 |
Anti-Hiv Neutralizing Antibody Ab1303 Fab Isolated From Sequentially Immunized Mcaques
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-01-19 Classification: IMMUNE SYSTEM |
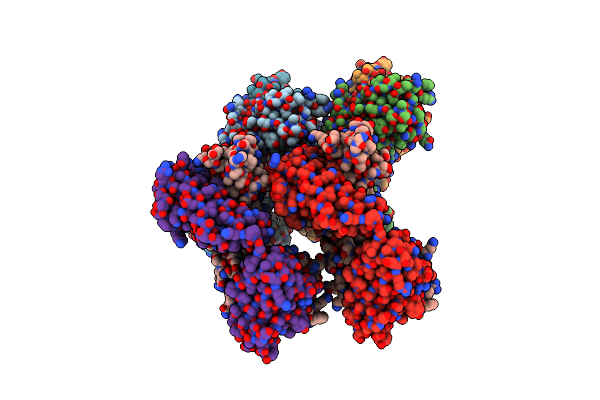 |
Anti-Hiv Neutralizing Antibody Ab1573 Fab Isolated From Sequentially Immunized Macaques
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-19 Classification: IMMUNE SYSTEM |
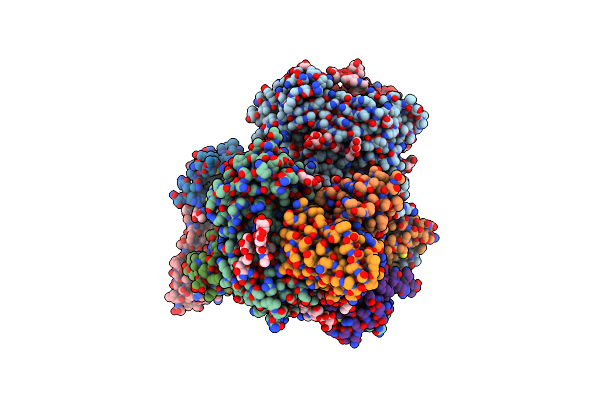 |
Cryo-Em Structure Of Cd4Bs Antibody Ab1303 In Complex With Hiv-1 Env Trimer Bg505 Sosip.664
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Resolution:4.00 Å Release Date: 2022-01-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, MAN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2018-09-19 Classification: membrane protein/inhibitor Ligands: CA, FJ7 |
 |
Organism: Arabidopsis thaliana, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2018-09-19 Classification: MEMBRANE PROTEIN Ligands: CA, PLM |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2018-09-19 Classification: MEMBRANE PROTEIN Ligands: CA, PLM, 3PH |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2018-09-19 Classification: MEMBRANE PROTEIN Ligands: CA, PLM, 3PH |
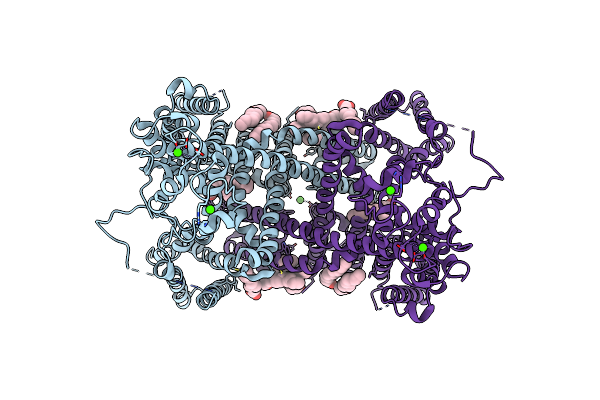 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2018-09-19 Classification: MEMBRANE PROTEIN Ligands: CA, PLM |
 |
Crystal Structure Of Rhodopsin Bound To Visual Arrestin Determined By X-Ray Free Electron Laser
Organism: Enterobacteria phage rb55, Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2017-08-09 Classification: SIGNALING PROTEIN |
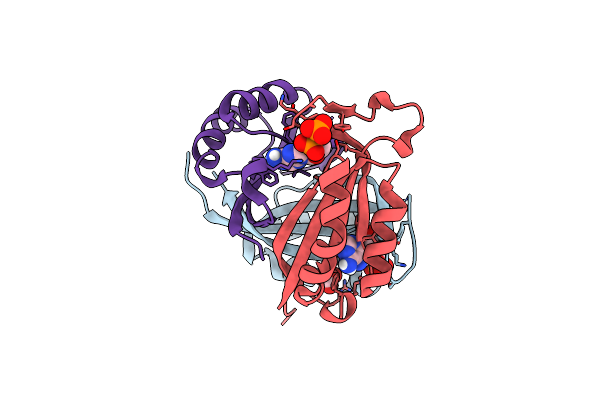 |
2.3 Angstrom Structure Of Cpii, A Nitrogen Regulatory Pii-Like Protein From Thiomonas Intermedia K12, Bound To Adp, Amp And Bicarbonate.
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2016-10-12 Classification: METAL BINDING PROTEIN Ligands: AMP, ADP, BCT |
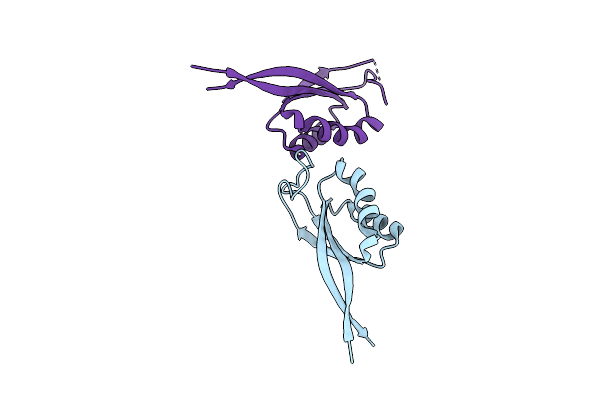 |
Structure Of The Apo Form Of Cpii From Thiomonas Intermedia K12, A Nitrogen Regulatory Pii-Like Protein
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-09-28 Classification: SIGNALING PROTEIN |
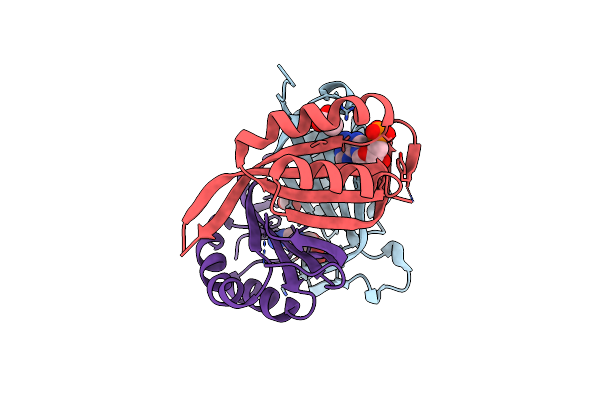 |
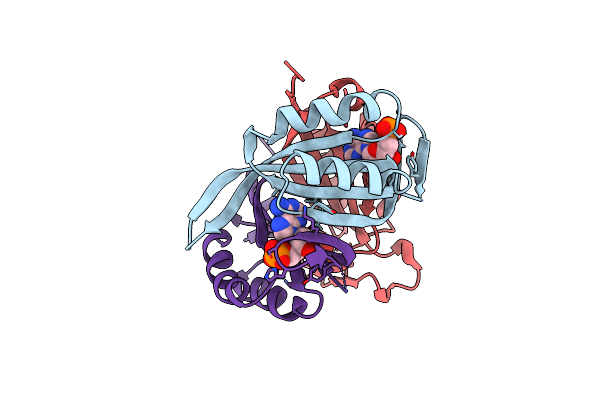
Structure Of Cpii Bound To Adp, Amp And Acetate, From Thiomonas Intermedia K12
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-09-28 Classification: SIGNALING PROTEIN Ligands: ADP, ACT, AMP |
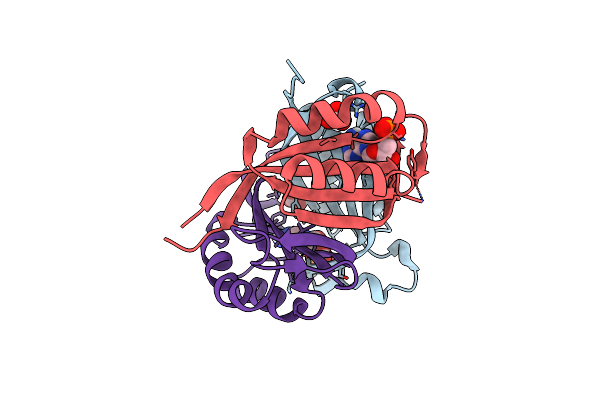 |
Structure Of Cpii, A Nitrogen Regulatory Pii-Like Protein From Thiomonas Intermedia K12, Bound To Adp, Amp And Bicarbonate.
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-28 Classification: SIGNALING PROTEIN Ligands: ADP, BCT, AMP |
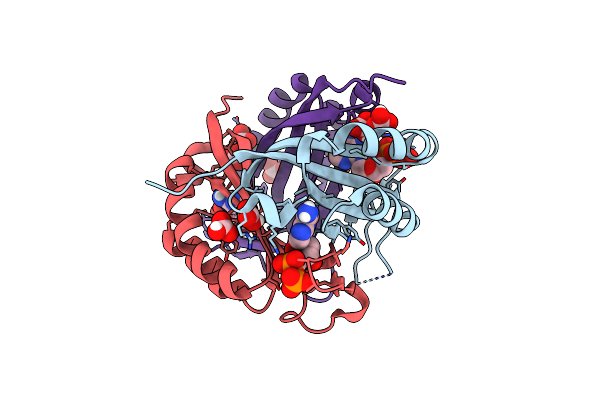 |
Structure Of Cpii Bound To Adp And Bicarbonate, From Thiomonas Intermedia K12
Organism: Thiomonas arsenitoxydans (strain dsm 22701 / cip 110005 / 3as)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-09-28 Classification: SIGNALING PROTEIN Ligands: ADP, BCT |
 |
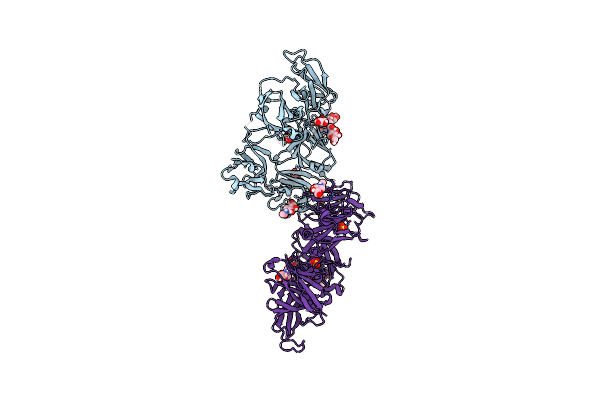
2.0 A Structure Of Cpii, A Nitrogen Regulatory Pii-Like Protein From Thiomonas Intermedia K12, Bound Amp
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-09-28 Classification: METAL BINDING PROTEIN Ligands: AMP, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-03-16 Classification: IMMUNE SYSTEM Ligands: NAG, SO4 |
 |
Crystal Structure Of The Teleost Fish Polymeric Ig Receptor (Pigr) Ectodomain
Organism: Oncorhynchus mykiss
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-03-16 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Rhodopsin Bound To Arrestin By Femtosecond X-Ray Laser
Organism: Enterobacteria phage t4, Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2015-07-29 Classification: SIGNALING PROTEIN |

