Search Count: 17
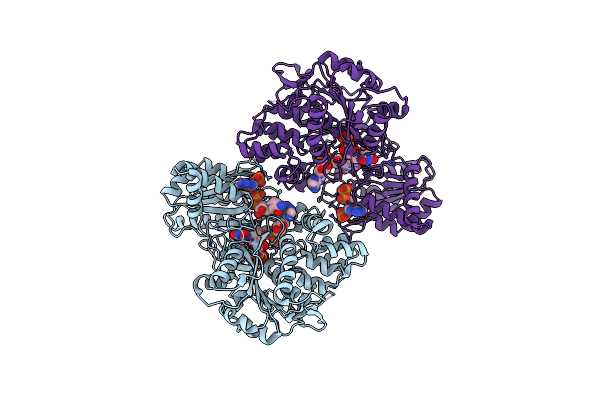 |
Crystal Structure Of A Tandem Deletion Mutant Of Rat Nadph-Cytochrome P450 Reductase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2021-01-13 Classification: FLAVOPROTEIN, OXIDOREDUCTASE Ligands: FMN, FAD, NAP |
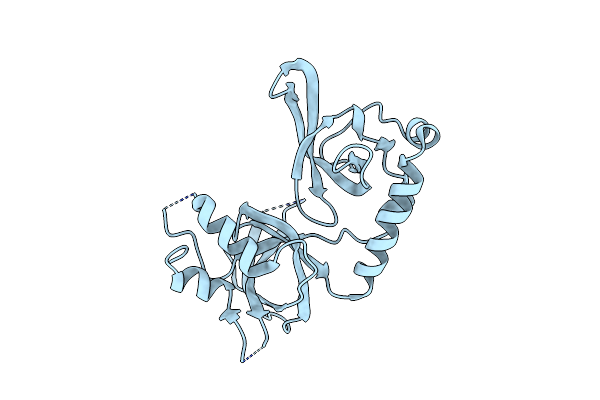 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-06-14 Classification: RNA BINDING PROTEIN |
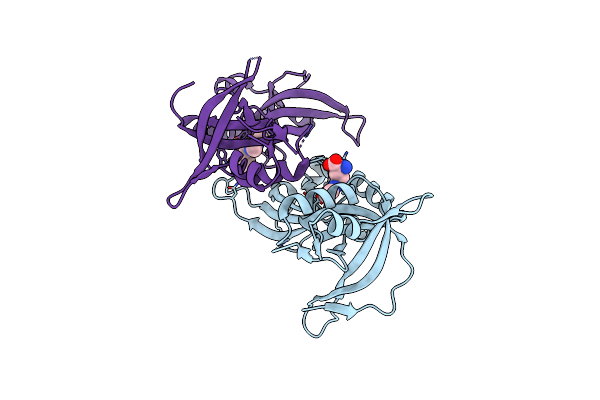 |
Crystal Structure Of The Extended Tudor Domain From Bmpapi In Complex With Sdma
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-06-14 Classification: RNA BINDING PROTEIN Ligands: 2MR |
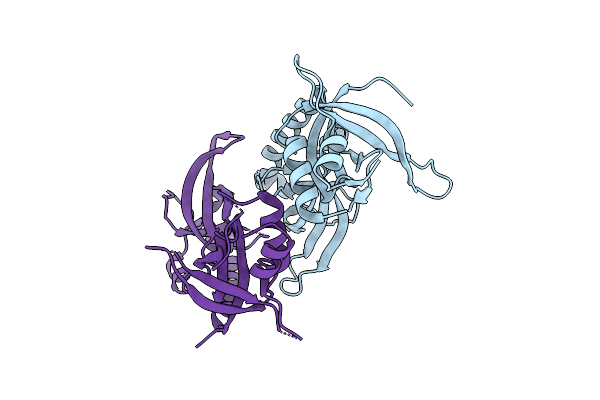 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2017-06-14 Classification: RNA BINDING PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-04-01 Classification: IMMUNE SYSTEM |
 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, And Mg (Holo-Icmf/Gdp)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: B12, 5AD, GDP, MG, ACT |
 |
Isobutyryl-Coa Mutase Fused With Bound Butyryl-Coa And Without Cobalamin Or Gdp (Apo-Icmf)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: BCO, TLA |
 |
Isobutyryl-Coa Mutase Fused With Bound Butyryl-Coa, Gdp, And Mg And Without Cobalamin (Apo-Icmf/Gdp)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: BCO, GDP, MG |
 |
Organism: Naegleria gruberi
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2013-10-02 Classification: TRANSFERASE |
 |
Structure Of A Eukaryotic Thiaminase-I Bound To The Thiamin Analogue 3-Deazathiamin
Organism: Naegleria gruberi
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2013-10-02 Classification: TRANSFERASE Ligands: 0YN |
 |
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-08-28 Classification: CHAPERONE Ligands: PO4, GDP |
 |
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-08-28 Classification: CHAPERONE Ligands: PO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-11-23 Classification: ISOMERASE Ligands: PO4, ZN, BEN |
 |
The Crystal Structure And Reaction Mechanism Of E. Coli 2,4-Dienoyl Coa Reductase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-09-30 Classification: OXIDOREDUCTASE Ligands: CL, SF4, FAD, FMN, NAP, MDE |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2001-08-22 Classification: OXIDOREDUCTASE Ligands: FAD, FMN, NAP |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2001-08-22 Classification: OXIDOREDUCTASE Ligands: FAD, FMN, NAP |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-08-22 Classification: OXIDOREDUCTASE Ligands: FAD, FMN, NAP, EPE |

