Search Count: 98
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Uq1-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, UQ1, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Pydiflumetofen-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, A1EE4, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Y19315-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, A1EKV, PEE |
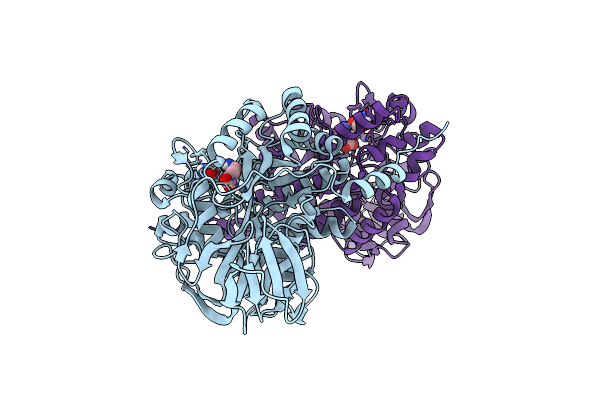 |
Crystal Structure Of Dihydropyrimidinase Complexed With Gamma-Aminobutyric Acid
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: ABU, ZN |
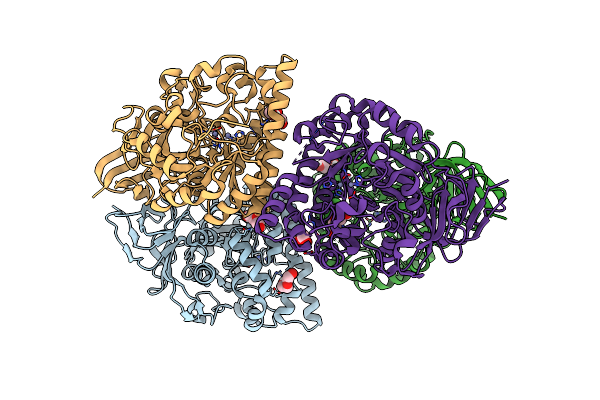 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-10-18 Classification: HYDROLASE Ligands: PEG, ZN |
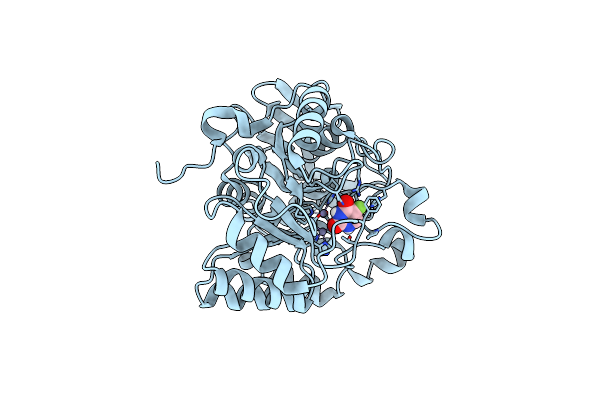 |
Crystal Structure Of The Human Dihydroorotase Domain In Complex With The Anticancer Drug 5-Fluorouracil
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-09-20 Classification: HYDROLASE Ligands: URF, ZN |
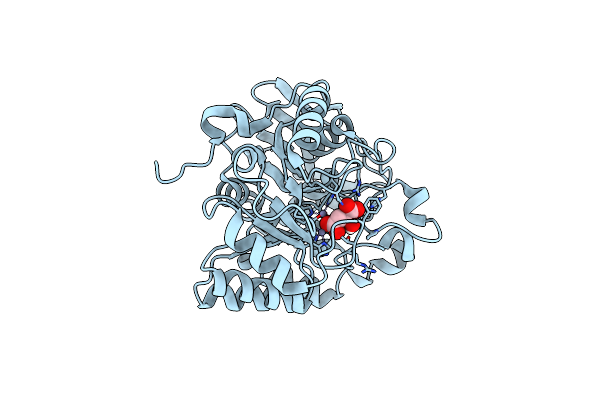 |
Crystal Structure Of The Human Dihydroorotase Domain In Complex With Malic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-09-20 Classification: HYDROLASE Ligands: LMR, ZN |
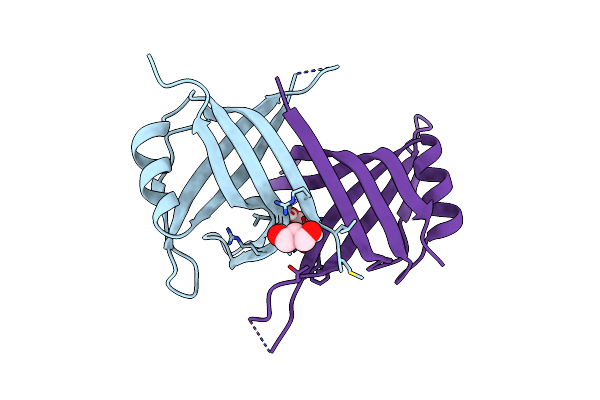 |
Organism: Staphylococcus aureus subsp. aureus ed98
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-20 Classification: DNA BINDING PROTEIN Ligands: GOL |
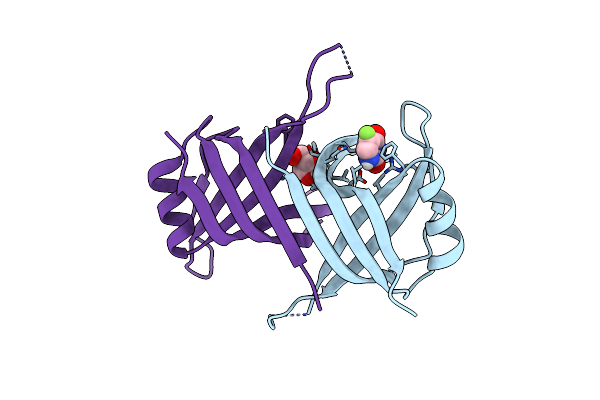 |
Structure Of Ssba Protein In Complex With The Anticancer Drug 5-Fluorouracil
Organism: Staphylococcus aureus subsp. aureus ed98
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-08-02 Classification: DNA BINDING PROTEIN Ligands: URF, GOL |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In D6-Dmso With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In D6-Dmso With Cis/Trans Switching (A-Cc Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
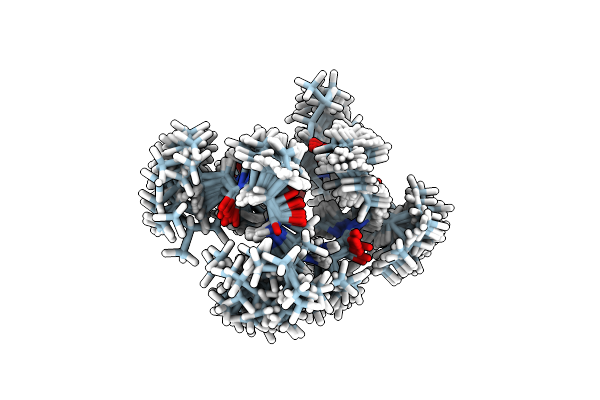 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In D6-Dmso With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
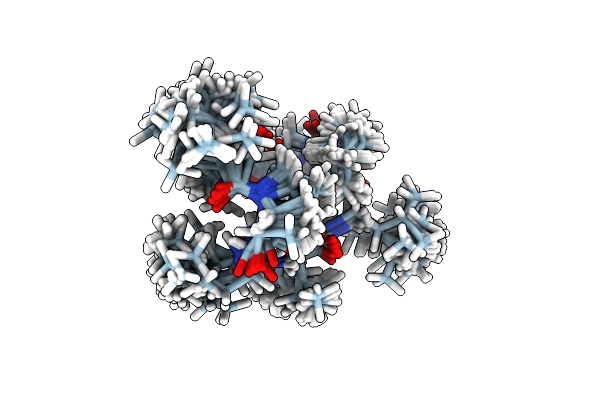 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In 50% D6-Dmso And 50% Water With Cis/Trans Switching (Cc Conformation, 50%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In Cdcl3 With Cis/Trans Switching (A-Tt Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In Cdcl3 With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In Cdcl3 With Cis/Trans Switching (Tt Conformation, 47%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2022-09-14 Classification: DE NOVO PROTEIN Ligands: HOH |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In Cdcl3 With Cis/Trans Switching (B-Tc Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In D6-Dmso With Cis/Trans Switching (B-Ct Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |

