Search Count: 467
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2026-01-14 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2026-01-14 Classification: NUCLEAR PROTEIN |
 |
Structure Of Bfl1 In Complex With A Covalent Inhibitor, Alternative Series, Cmpd25
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: APOPTOSIS Ligands: A1JL2 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: G6P |
 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: A1EUP |
 |
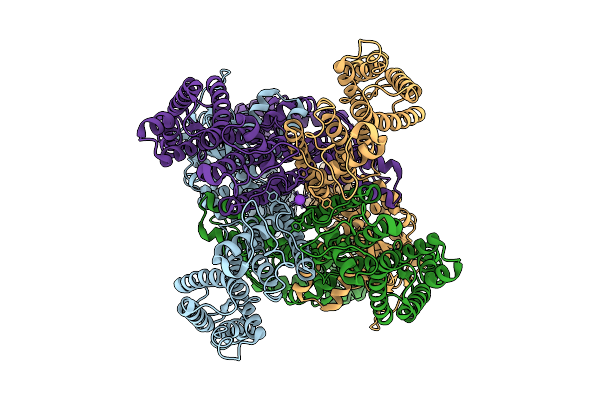
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: K |
 |
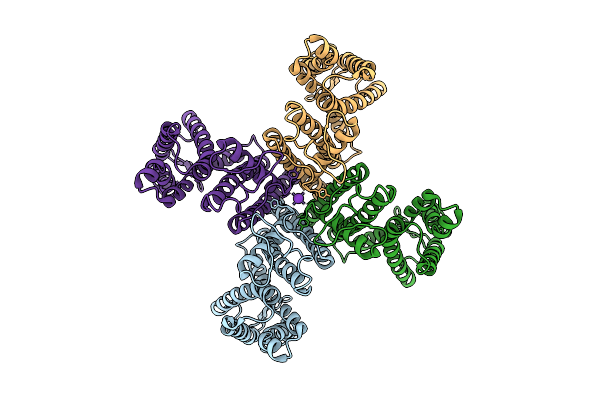
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTON TRANSPORT Ligands: K |
 |
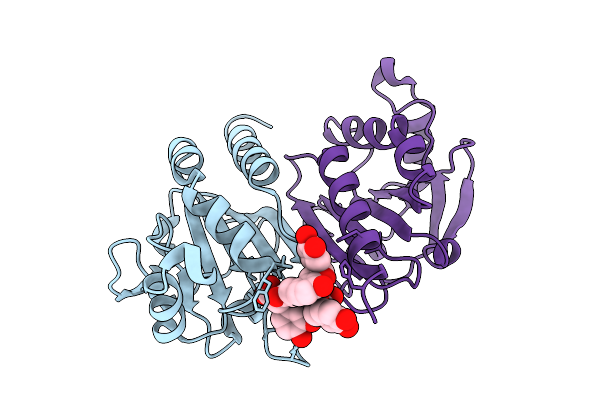
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTON TRANSPORT Ligands: K |
 |
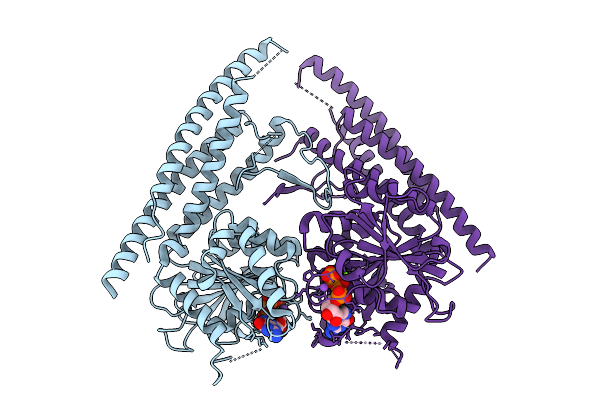
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: OXIDOREDUCTASE Ligands: A1LXC |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GTP, MG, NA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GTP, MG, K |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GDP, BEF, MG, K, MES |
 |
Organism: Echinococcus multilocularis
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE |
 |
Organism: Spodoptera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: AND |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: PROTEIN BINDING Ligands: ZN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: PROTEIN BINDING Ligands: ZN |

