Search Count: 22
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, PO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, PO4, K |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, PO4, K |
 |
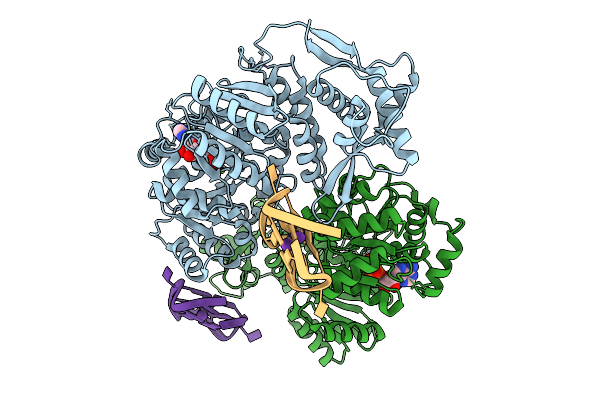
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, K, PGE |
 |
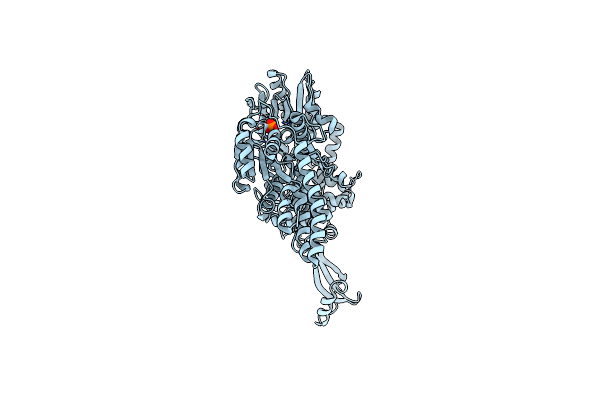
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, K |
 |
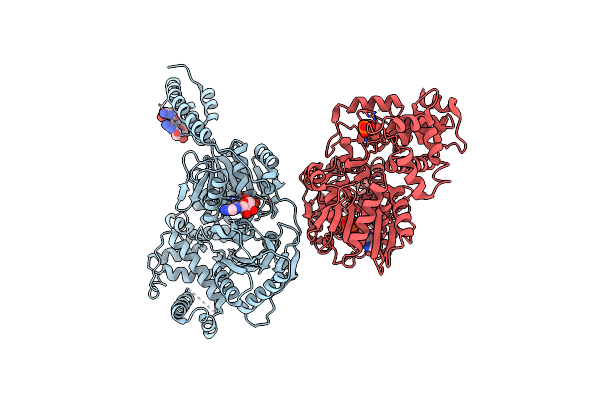
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: PO4 |
 |
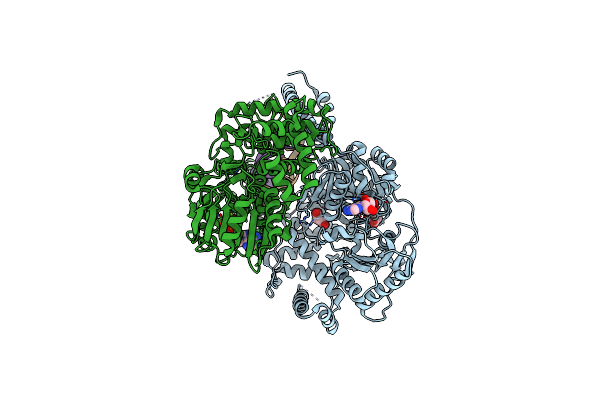
Structure Of Escherichia Coli Hrpa In Complex With Adp And Dinucleotide Dcdc
Organism: Escherichia coli k-12, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: ADP, MG, PO4 |
 |
Structure Of Escherichia Coli Hrpa In Complex With Adp And Oligonucleotide Poly(Dc)11 Forming An I-Motif
Organism: Escherichia coli k-12, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: ADP, MG, SIN |
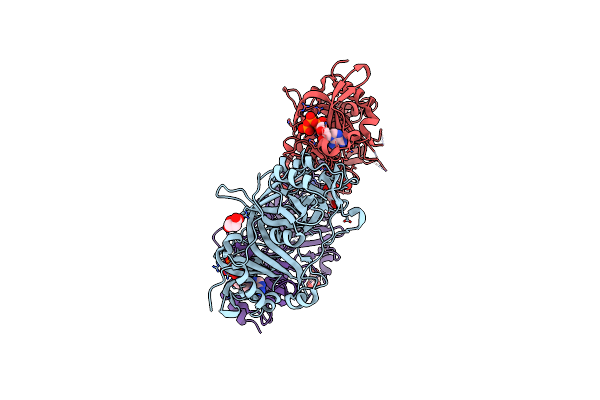 |
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Release Date: 2024-04-17 Classification: PLANT PROTEIN Ligands: ATP, MG, SIN |
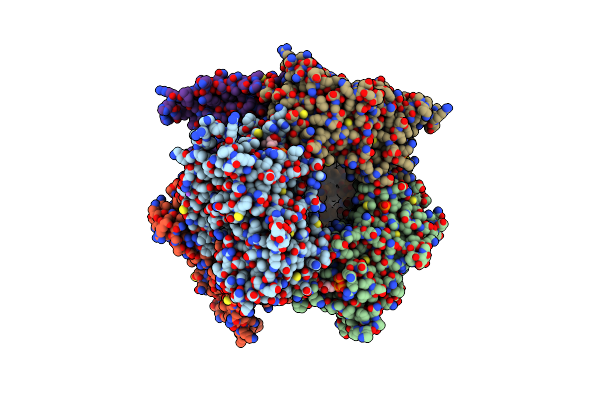 |
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2024-04-17 Classification: PLANT PROTEIN Ligands: ATP, MG, PO4, NA |
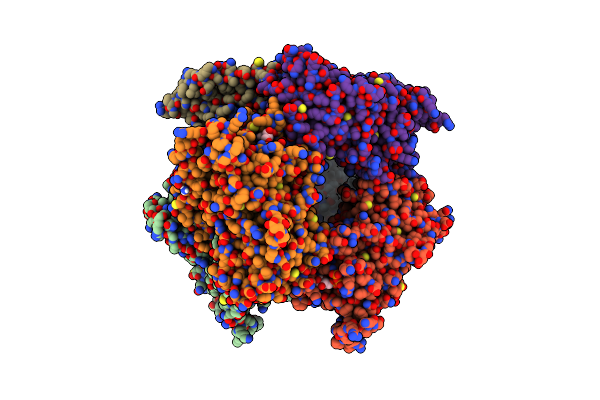 |
Structure Of Nal1 Indica Cultivar Ir64, Construct 36-458 In Presence Of Peptide From Fzp Protein
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2024-04-17 Classification: PLANT PROTEIN Ligands: ATP, MG, SO4, NH4 |
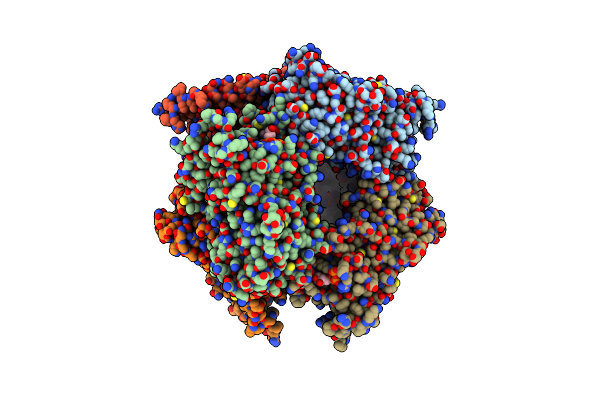 |
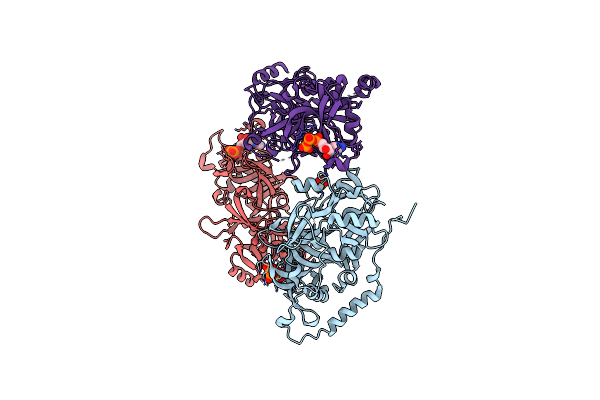
Structure Of Nal1 Protein , Spike Allele From Japonica Rice, Construct 46-458
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2024-04-17 Classification: PLANT PROTEIN Ligands: ATP, MG |
 |
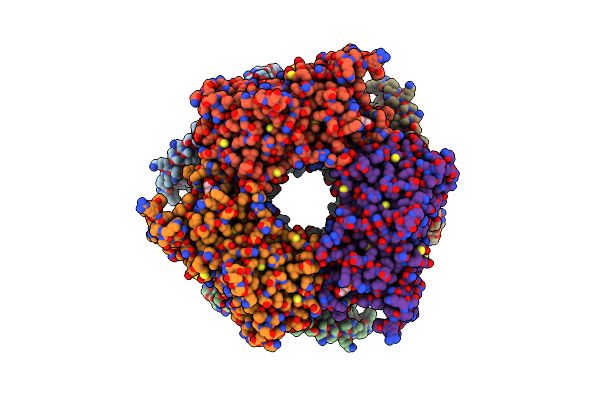
Structure Of Nal1 Protein, Allele Spike From Japonica Rice, Construct 31-458
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-17 Classification: PLANT PROTEIN Ligands: ATP, MG, PO4 |
 |
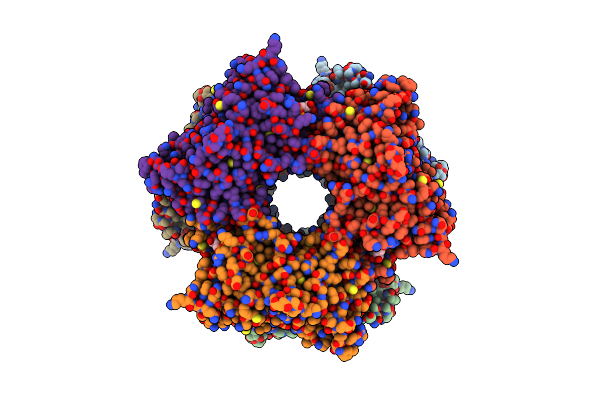
Cryoem Structure Of Nal1 Protein, Allele Spike, From Oryza Sativa Japonica Group
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PLANT PROTEIN Ligands: ATP, MG |
 |
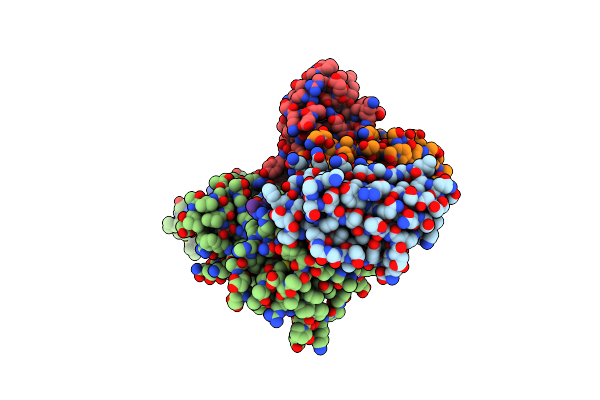
Cryoem Structure Of Nal1 Protein, Allele Ir64, From Oryza Sativa Indica Cultivar
Organism: Oryza sativa indica group
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PLANT PROTEIN Ligands: ATP, MG |
 |
Stabilized Beta-Arrestin 1-V2T Subcomplex Of A Gpcr-G Protein-Beta-Arrestin Mega-Complex
Organism: Lama glama, Bos taurus, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2019-11-20 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Enterobacteria phage rb59, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2019-11-20 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: P0G |
 |
Structure Of Beta2 Adrenergic Receptor Bound To Bi167107, Nanobody 6B9, And A Positive Allosteric Modulator
Organism: Enterobacteria phage rb59, Homo sapiens, Camelidae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-06-26 Classification: SIGNALING PROTEIN Ligands: P0G, 1WV, KBY |
 |
Organism: Triticum turgidum
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2017-10-25 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-08-16 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SQZ |

