Search Count: 548
 |
Organism: Thermothielavioides terrestris (strain atcc 38088 / nrrl 8126)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, MG |
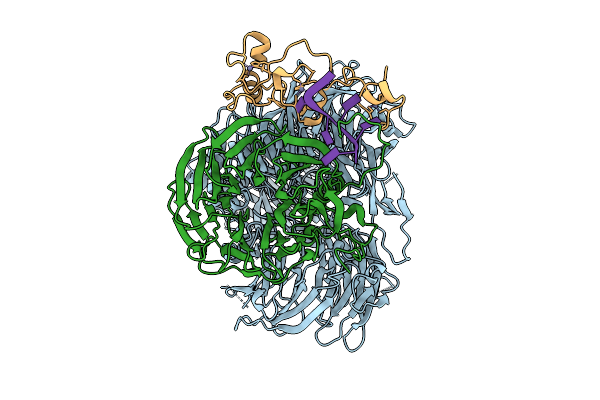 |
Cryo-Em Structure Of The Human Mpsf In Complex With The Aguaaa Poly(A) Signal
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
 |
Cryo-Em Structure Of The Human Mpsf In Complex With The Aauaau Poly(A) Signal
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
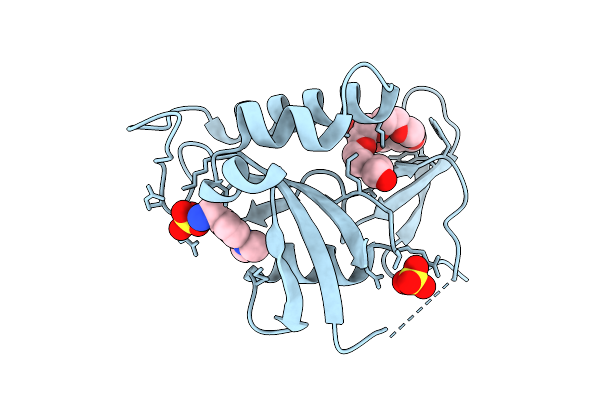 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: ISOMERASE Ligands: A1ED1, PE8, SO4 |
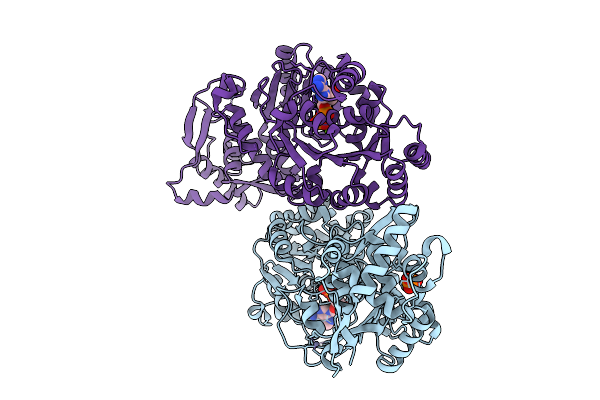 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, PO4 |
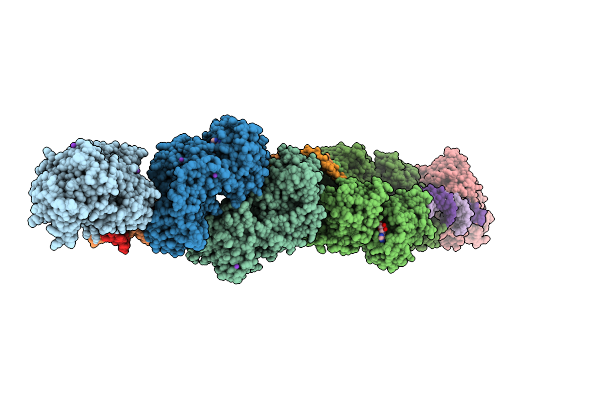 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, PO4, K |
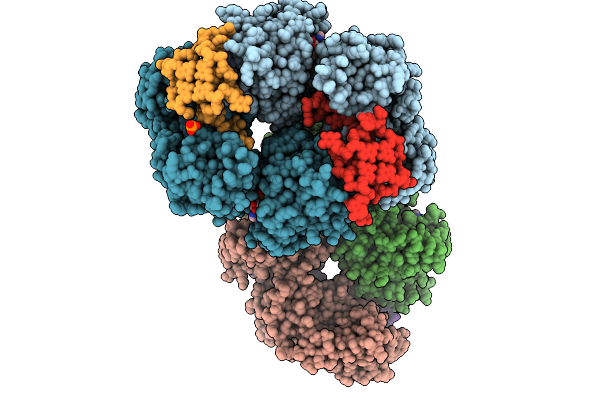 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, PO4, K |
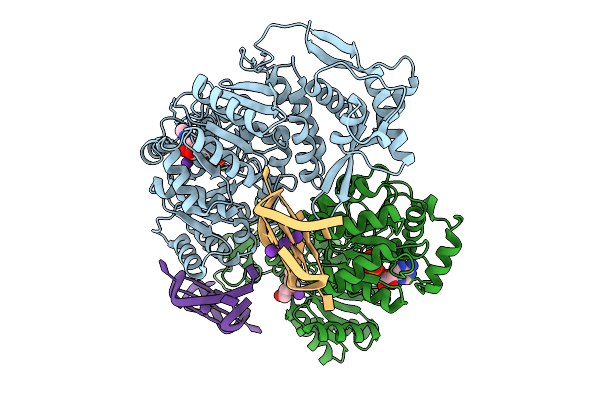 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, K, PGE |
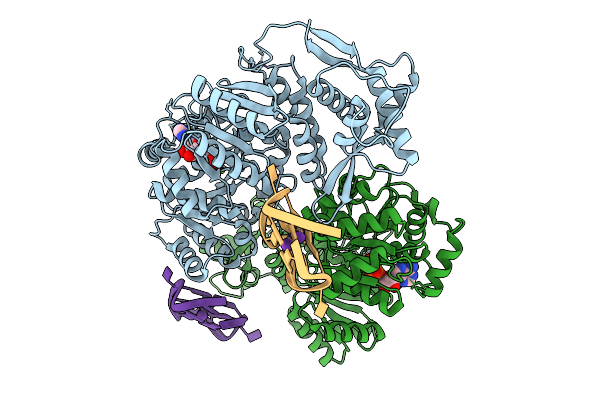 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ZN, ADP, MG, K |
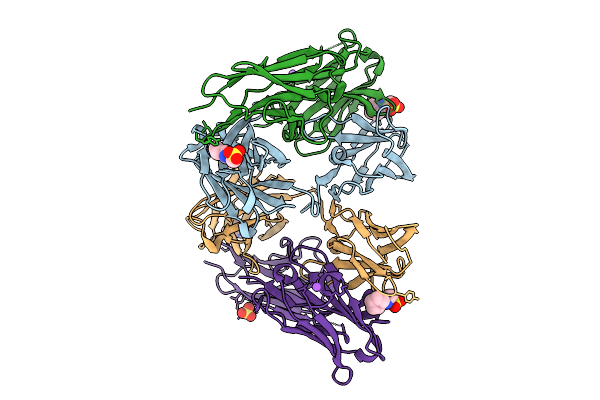 |
Crystal Structure Of A High Affinity Vl-Vh Tetrabody For The Erythropoietin Receptor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: SIGNALING PROTEIN Ligands: NHE, CL, SO4, NA |
 |
Crystal Structure Of The Methyltransferase-Ribozyme 1 Bound To Dna Substrate (With 1-Methyl-Adenosine)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DNA-RNA HYBRID |
 |
Crystal Structure Of The Methyltransferase-Ribozyme 1 Bound To Dna Substrate (1-Benzyl-Adenosine Derivative)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DNA-RNA HYBRID |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: RNA Ligands: MG |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: RNA Ligands: MG, K |
 |
Organism: Phocaeicola dorei cl03t12c01
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: RNA Ligands: SO4, NA |
 |
Organism: Micromonospora echinaurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Micromonospora echinaurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE |

