Search Count: 26
 |
Acarbose Hydrolase From Human Gut Microbiota K. Grimontii Td1, Apg Mutant Enzyme D336A, Complexed With Acarviosine-Glucose
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE |
 |
Apg Mutant Enzyme D336A Of Acarbose Hydrolase From Human Gut Flora K. Grimontii Td1, Complex With Acarviosine
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: A1EAX |
 |
Apg Mutant Enzyme D448A Of Acarbose Hydrolase From Human Gut Flora K. Grimontii Td1, Complex With Acarbose
Organism: Klebsiella grimontii, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE |
 |
Apg Mutant Enzyme D448A Of The Human Gut Flora K. Grimontii Td1 Acarbose Hydrolase
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE |
 |
Apg, Crystal Structure Of Acarbose Hydrolase From The Human Gut Flora K. Grimontii Td1
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE |
 |
Cryo-Em Structure Of Human 26S Rp (Ed State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Four Ub.
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Human 26S Rp (Eb State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Four Ub.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Human 26S Proteasomal Rp Subcomplex (Ea State) Without Any Bound Substrate.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Human 26S Proteasomal Rp Subcomplex (Ea State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Three Ub.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: PROTEIN FIBRIL |
 |
Cryoem Structure Of Human Alpha-Synuclein A53T Fibril Induced By Calcium Ions
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: PROTEIN FIBRIL |
 |
Organism: Mesocricetus auratus
Method: ELECTRON MICROSCOPY Release Date: 2022-07-27 Classification: PROTEIN FIBRIL |
 |
 |
Organism: Thermococcus sp. 9on-7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-08-28 Classification: TRANSFERASE/DNA Ligands: CA |
 |
Organism: Thermococcus sp. 9on-7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-08-28 Classification: TRANSFERASE/DNA Ligands: CA |
 |
Organism: Thermococcus sp. 9on-7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2019-08-28 Classification: TRANSFERASE/DNA Ligands: PPV, CA |
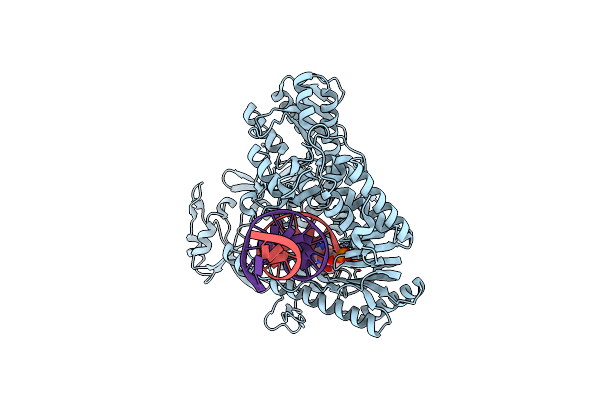 |
Structure Of 9N-I Dna Polymerase Incorporation With 3'-Al In The Active Site
Organism: Thermococcus sp. 9on-7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-08-28 Classification: TRANSFERASE/DNA Ligands: PPV, CA, B9X |
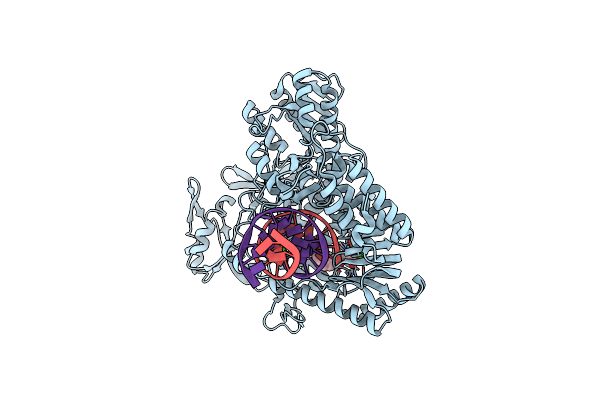 |
Structure Of 9N-I Dna Polymerase Incorporation With 3'-Cl In The Active Site
Organism: Thermococcus sp. 9on-7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-08-28 Classification: TRANSFERASE/DNA Ligands: PPV, CA, B0U |
 |
Organism: Vibrio parahaemolyticus m0605
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2015-08-26 Classification: TOXIN Ligands: NO3 |
 |
Organism: Vibrio parahaemolyticus m0605
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-08-26 Classification: TOXIN |

