Search Count: 50
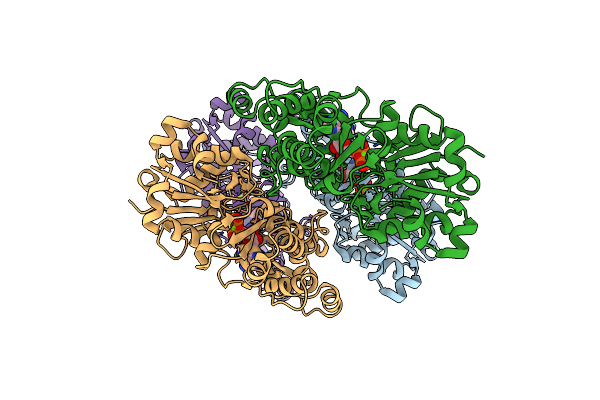 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.12 Å Release Date: 2025-05-14 Classification: STRUCTURAL PROTEIN Ligands: MG, ADP |
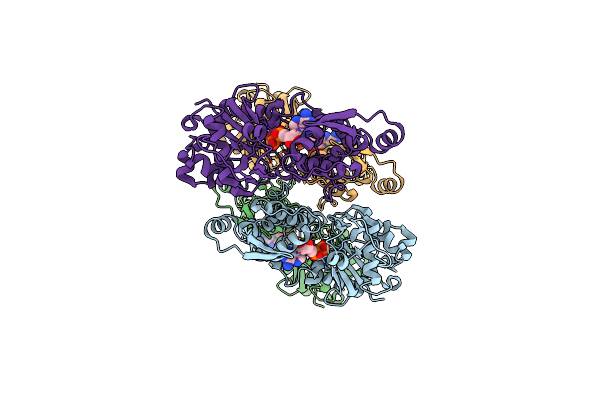 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: STRUCTURAL PROTEIN Ligands: ADP, MG |
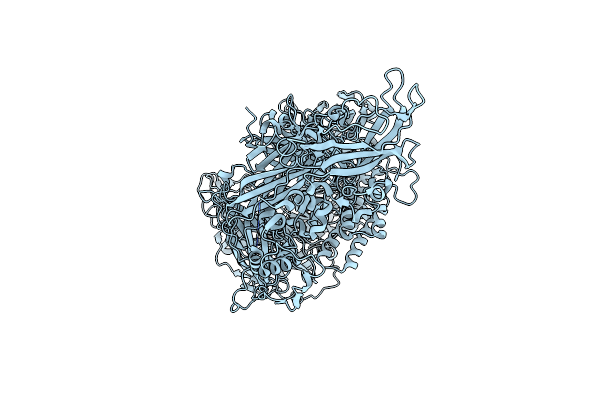 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: MEMBRANE PROTEIN |
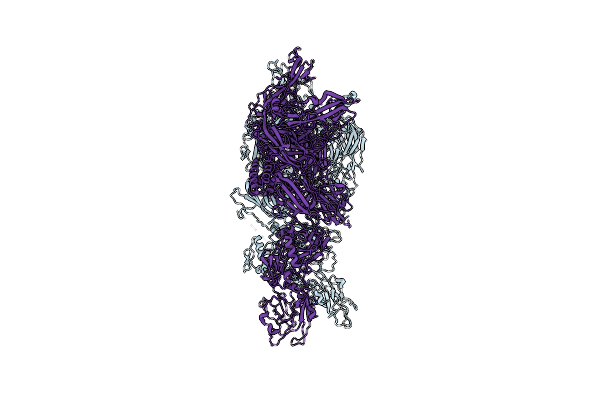 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: MEMBRANE PROTEIN |
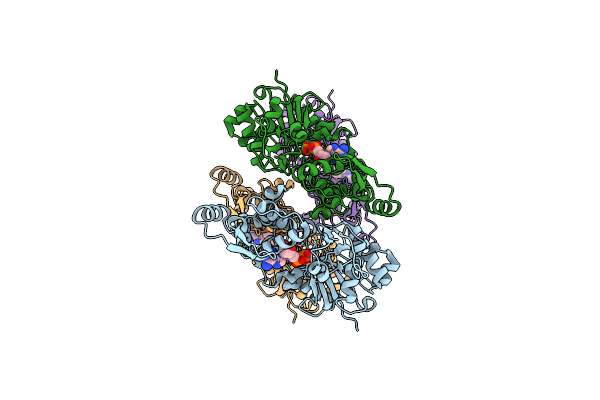 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: STRUCTURAL PROTEIN Ligands: MG, ADP |
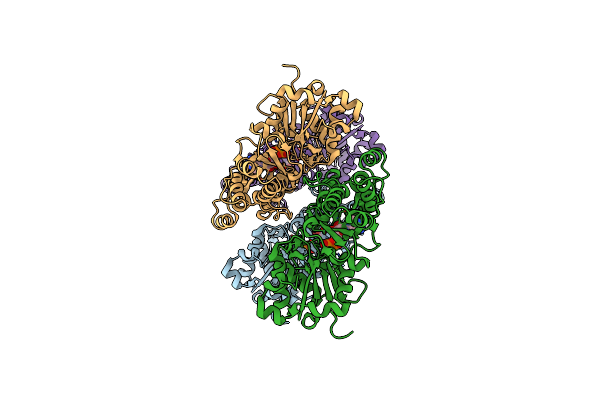 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: STRUCTURAL PROTEIN Ligands: ADP, MG |
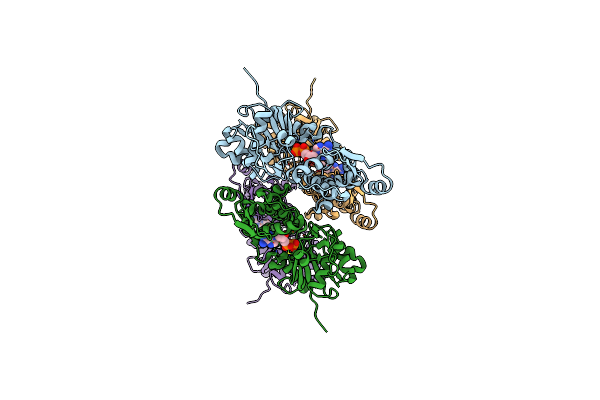 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: STRUCTURAL PROTEIN Ligands: MG, ADP |
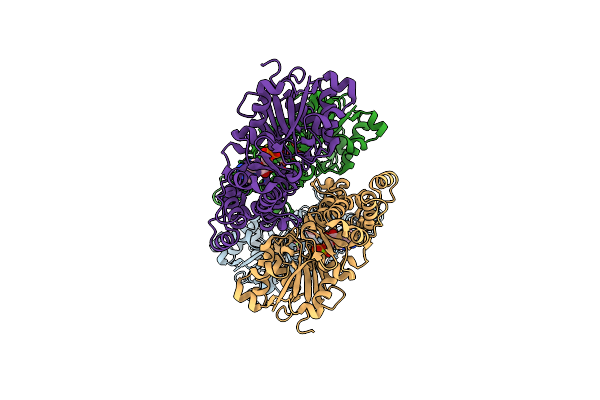 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: STRUCTURAL PROTEIN Ligands: MG, ADP |
 |
High-Resolution Structure Of Photosystem Ii From The Mesophilic Cyanobacterium, Synechocystis Sp. Pcc 6803
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: ELECTRON MICROSCOPY Release Date: 2021-12-29 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, CL, CLA, PHO, BCR, LMG, PL9, SQD, LMT, BCT, LHG, DGD, HEM, CA, RRX |
 |
High-Resolution Structure Of Photosystem Ii From The Mesophilic Cyanobacterium, Synechocystis Sp. Pcc 6803
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: ELECTRON MICROSCOPY Release Date: 2021-12-29 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, CL, CLA, PHO, BCR, SQD, LMG, PL9, LMT, BCT, LHG, DGD, HEM, CA, RRX |
 |
Quantitative Assessment Of Chlorophyll Types In Cryo-Em Maps Of Photosystem I Acclimated To Far-Red Light
Organism: Fischerella thermalis pcc 7521
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, F6C, PQN, SF4, BCR, LHG, LMG, LMT, CA |
 |
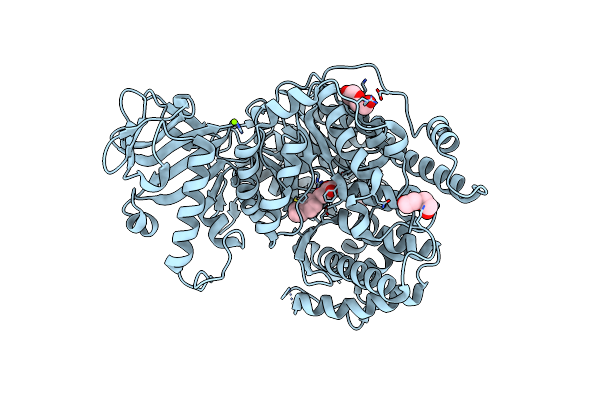
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With Dioxine-Phenyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-05 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: MG, BXS, PEG |
 |
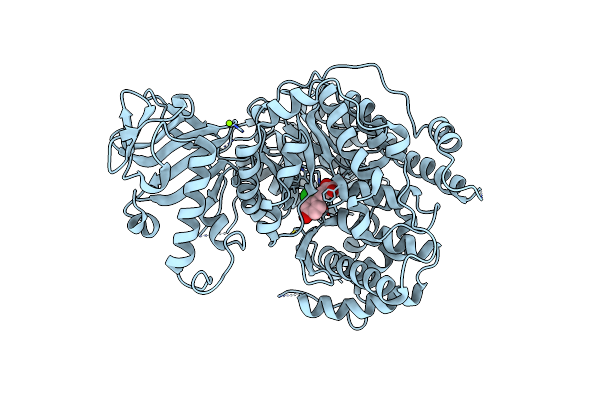
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With 2-Naphthyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-05 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: MG, C0V, PEG |
 |
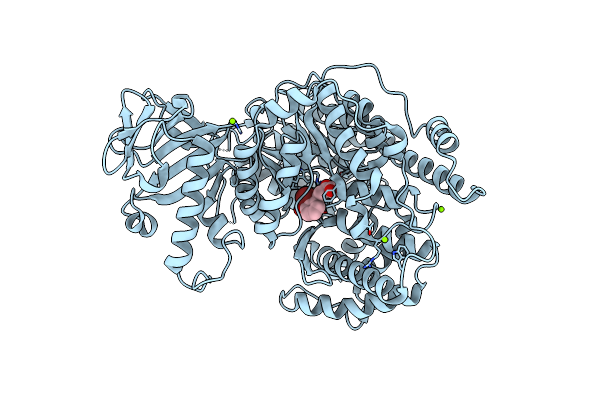
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With 2-Cl-4-Oh-Phenyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-09-05 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: MG, D1Y |
 |
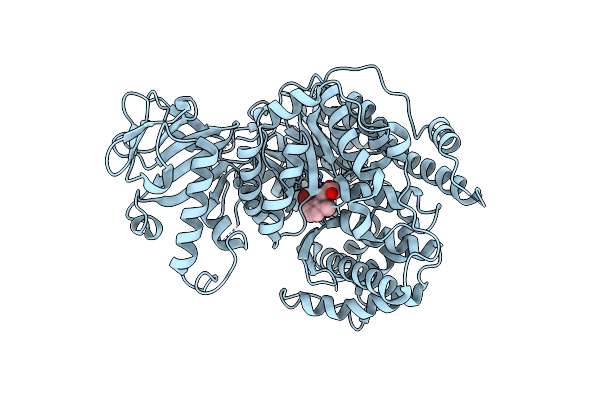
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With 2-Br-3-Oh-Phenyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2018-09-05 Classification: STRUCTURAL GENOMICS,Transferase Ligands: MG, E9S |
 |
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With 2-Br-6-Me-Phenyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-09-05 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: MG, EHV |
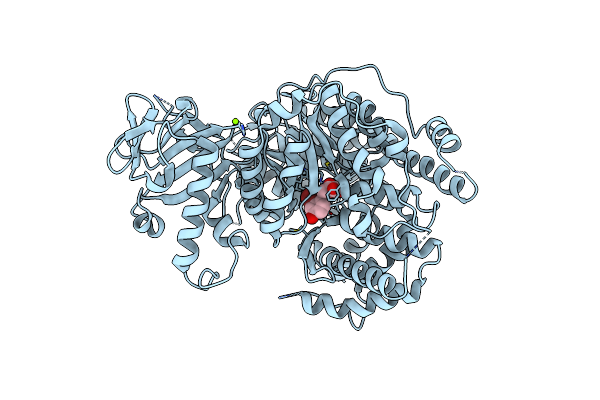 |
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With 2-Br-4-Oh-Phenyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-09-05 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: MG, ENG |
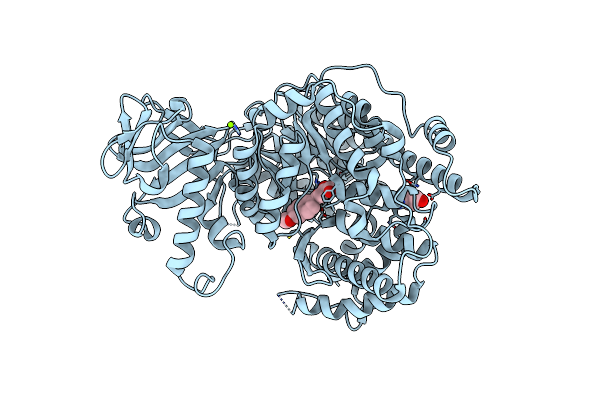 |
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With Methoxynaphthyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2018-09-05 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: ENY, MG, PEG |
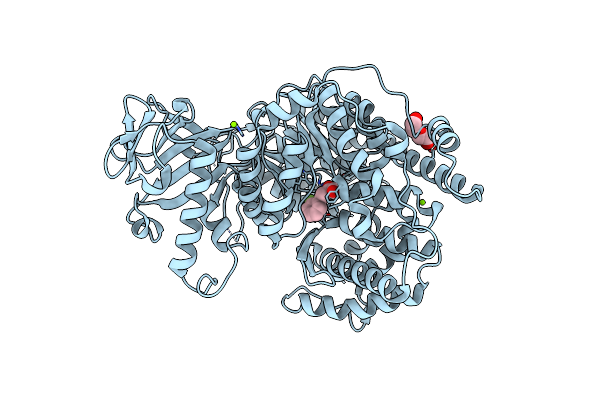 |
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With 2-F-Phenyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-09-05 Classification: TRANSFERASE Ligands: MG, EQA, PEG |
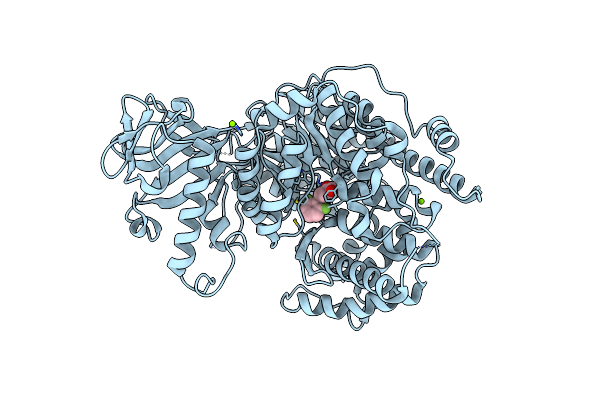 |
Crystal Structure Of Mycobacterium Tuberculosis Malate Synthase In Complex With 2,6-F-Phenyldiketoacid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2018-09-05 Classification: LYASE/LYASE inhibitor Ligands: MG, GXG |

