Search Count: 380
 |
Organism: Homo sapiens
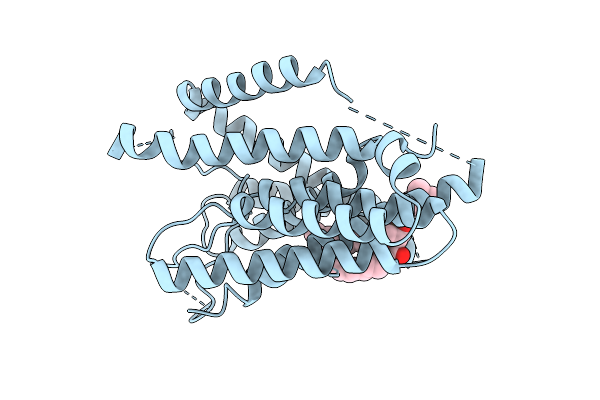
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: PC1 |
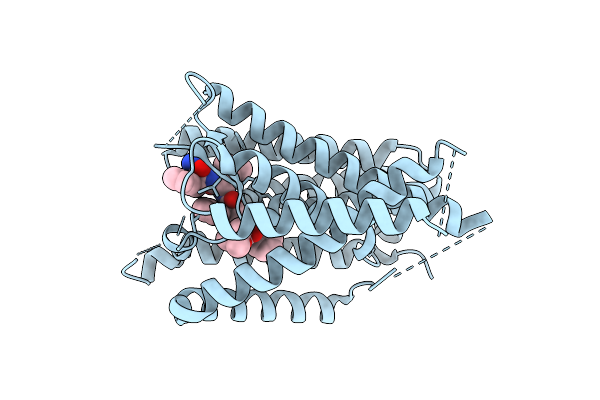 |
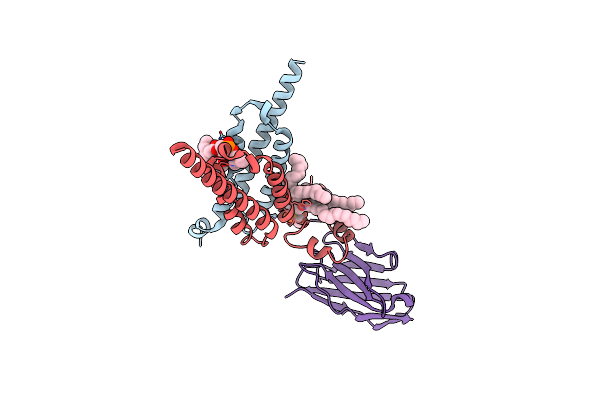
Cryo-Em Structure Of Human Signal Peptide Peptidase Like 2A (Sppl2A) In Complex With L685,458
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: FTO |
 |
Organism: Homo sapiens
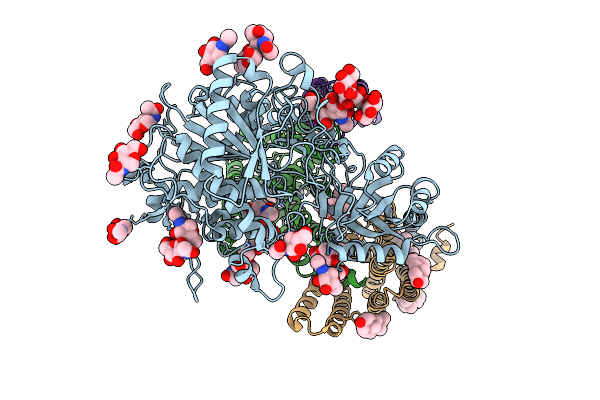
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: NAG, PC1, A1D6X, CLR |
 |
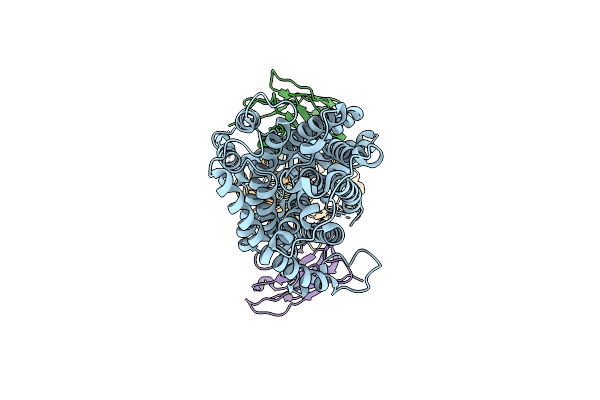
Organism: Arabidopsis thaliana
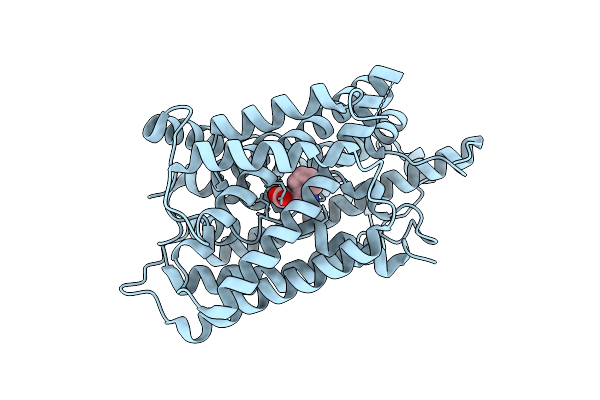
Method: ELECTRON MICROSCOPY Resolution:3.52 Å Release Date: 2025-07-23 Classification: PROTEIN TRANSPORT Ligands: IAC |
 |
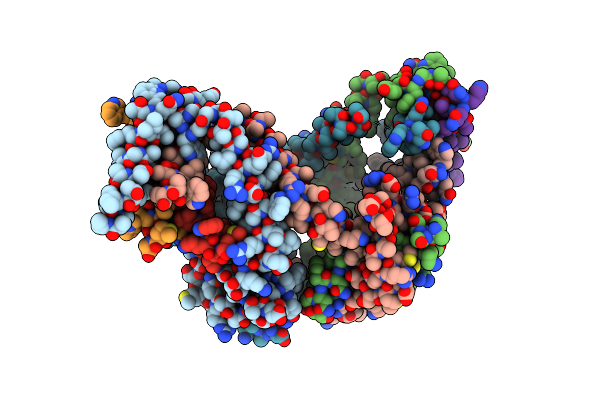
Organism: Arabidopsis thaliana, Escherichia coli, Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:3.97 Å Release Date: 2025-07-23 Classification: PROTEIN TRANSPORT/IMMUNE SYSTEM |
 |
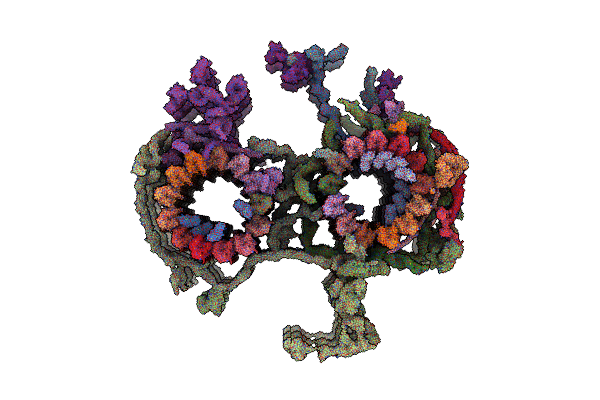
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: APOPTOSIS |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PROTEIN TRANSPORT |
 |
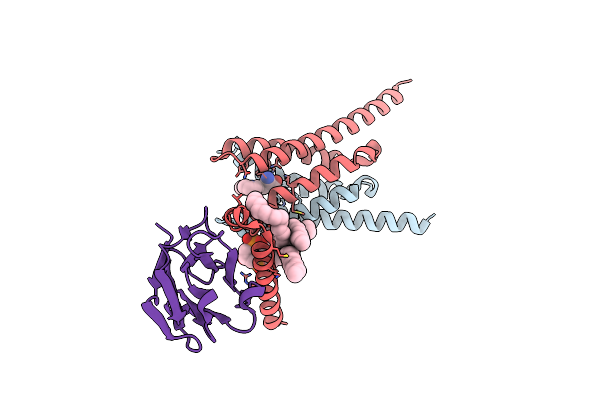
Cryo-Em Structure Of Apo Human Mitochondrial Pyruvate Carrier In The Ims-Open Conformation At Ph 8.0
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: PC8, CDL |
 |
Cryo-Em Structure Of Human Mitochondrial Pyruvate Carrier In Complex With The Inhibitor Uk5099
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL, I2R |
 |
Cryo-Em Structure Of Human Mitochondrial Pyruvate Carrier In The Matrix-Facing Conformation At Ph 6.8
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL |
 |
Cryo-Em Structure Of Apo Human Mitochondrial Pyruvate Carrier In The Ims-Open Conformation At Ph 6.8
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL, PC8 |
 |
Cryo-Em Structure Of Human Mitochondrial Pyruvate Carrier In The Occluded Conformation At Ph 6.8
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL |
 |
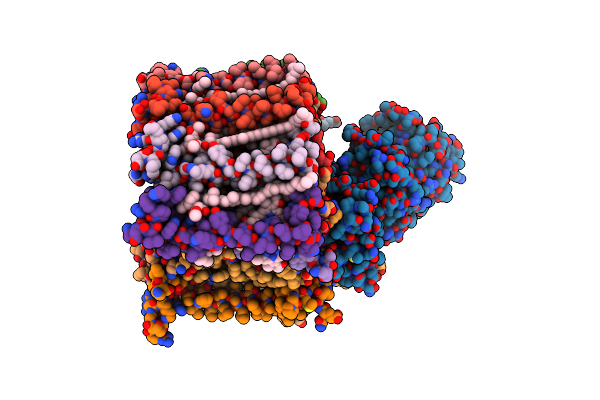
Cryo-Em Structure Of Pyruvate-Treated Human Mitochondrial Pyruvate Carrier In The Ims-Open Conformation At Ph 8.0
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL, PC8 |
 |
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Resolution:3.05 Å Release Date: 2025-02-19 Classification: PHOTOSYNTHESIS Ligands: BCL, U4Z, HEM, PGV, MQE, BPH, MN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN Ligands: NAG, CLR, ZN, P5S, CA |
 |
Organism: Artificial sequences
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-27 Classification: DE NOVO PROTEIN Ligands: KY6 |
 |
Organism: Artificial sequences
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-11-27 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial sequences, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: KY6 |
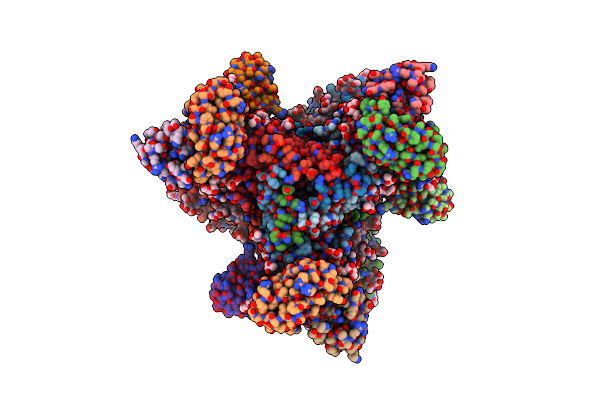 |
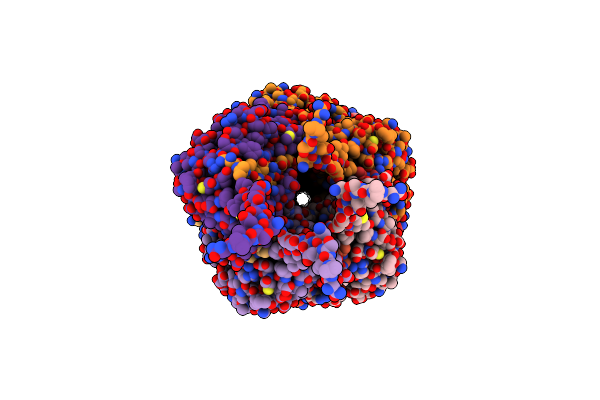
Structure Of Vrc44.01 Fab In Complex With 3Bnc117-Purified C1080.C3 Rns Sosip.664 Hiv-1 Env Trimer
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, PO4 |
 |
Organism: Algae metagenome
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: PROTEIN FIBRIL |

