Search Count: 3
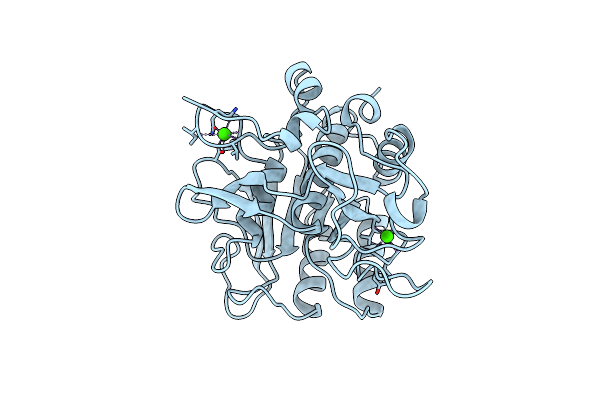 |
Subtilisin Bpn' 8397+1 (E.C. 3.4.21.14) (Mutant With Met 50 Replaced By Phe, Asn 76 Replaced By Asp, Gly 169 Replaced By Ala, Gln 206 Replaced By Cys, Asn 218 Replaced By Ser And Lys 256 Replaced By Tyr) (M50F, N76D, G169A, Q206C, N218S, And K256Y) In 20% Dimethylformamide
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1996-07-11 Classification: HYDROLASE (SERINE PROTEASE) Ligands: CA |
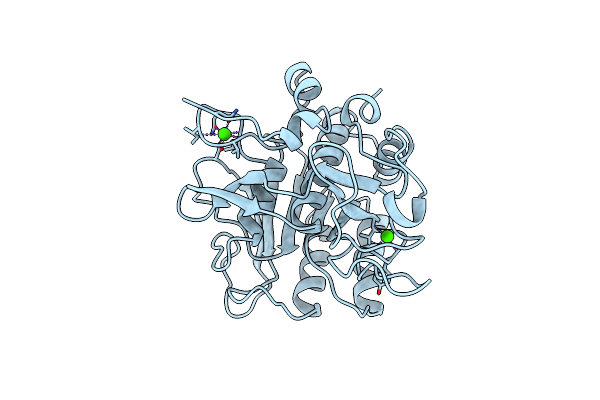 |
Subtilisin Bpn' 8397+1 (E.C. 3.4.21.14) (Mutant With Met 50 Replaced By Phe, Asn 76 Replaced By Asp, Gly 169 Replaced By Ala, Gln 206 Replaced By Cys, Asn 218 Replaced By Ser And Lys 256 Replaced By Tyr) (M50F, N76D, G169A, Q206C, N218S, And K256Y) In 35% Dimethylformamide
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1996-07-11 Classification: HYDROLASE (SERINE PROTEASE) Ligands: CA |
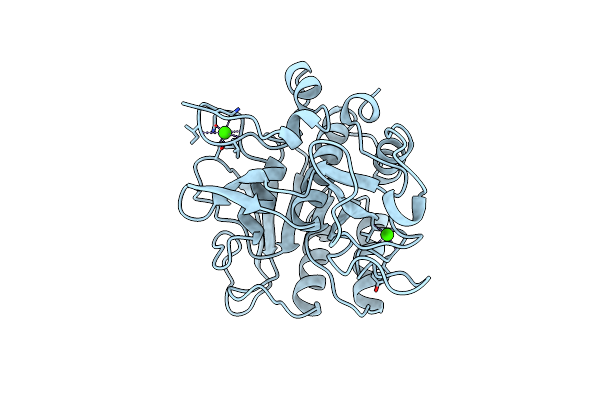 |
Subtilisin Bpn' 8397+1 (E.C. 3.4.21.14) (Mutant With Met 50 Replaced By Phe, Asn 76 Replaced By Asp, Gly 169 Replaced By Ala, Gln 206 Replaced By Cys, Asn 218 Replaced By Ser And Lys 256 Replaced By Tyr) (M50F, N76D, G169A, Q206C, N218S, And K256Y) In 50% Dimethylformamide
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1996-07-11 Classification: HYDROLASE (SERINE PROTEASE) Ligands: CA |

