Search Count: 919
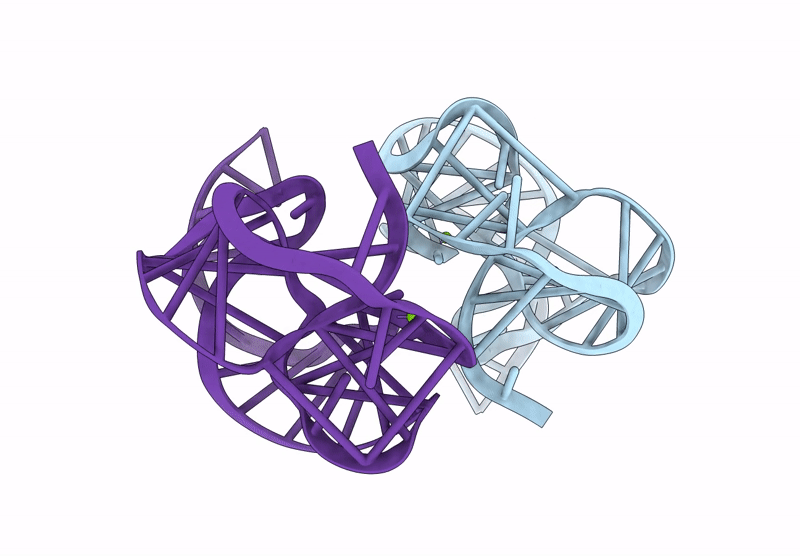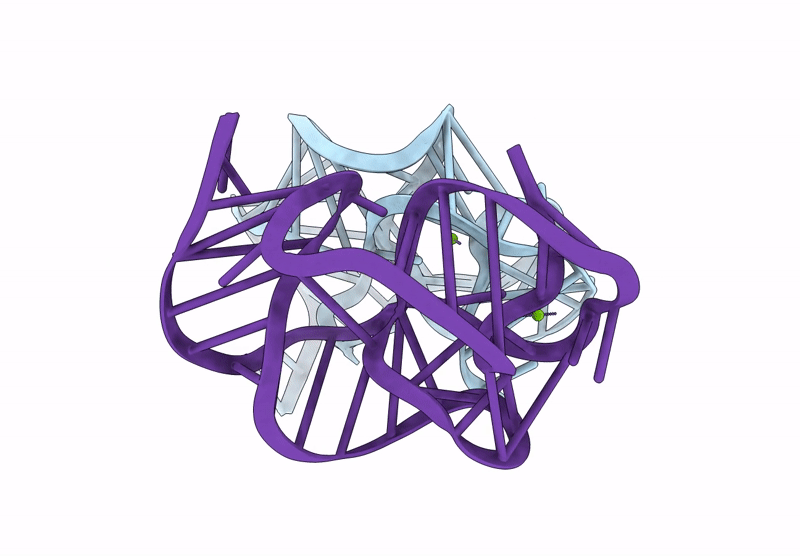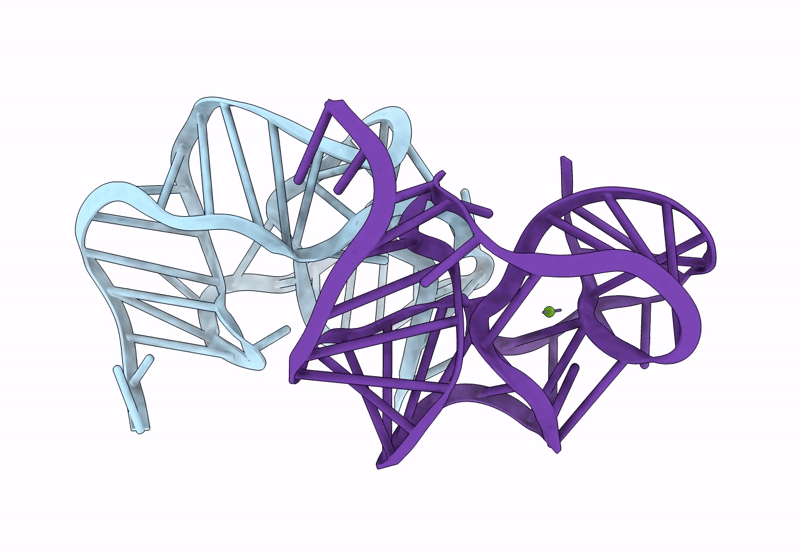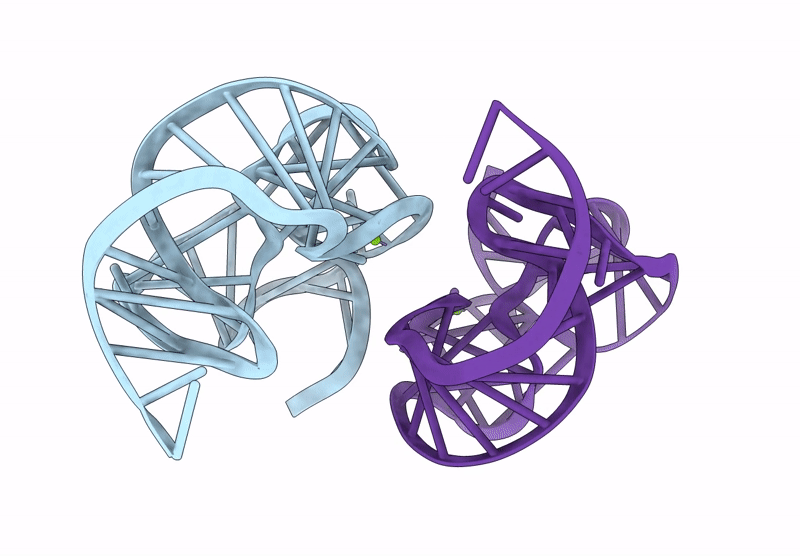 |
Crystal Structure Of An Exoribonuclease-Resistant Rna From A Tombusvirus-Like Associated Rna
Organism: Beet western yellows st9 associated virus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: RNA Ligands: IRI, MG |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LIGASE Ligands: GFD, A1EMI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LIGASE Ligands: A1EMN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LIGASE Ligands: GFD, A1EMM |
 |
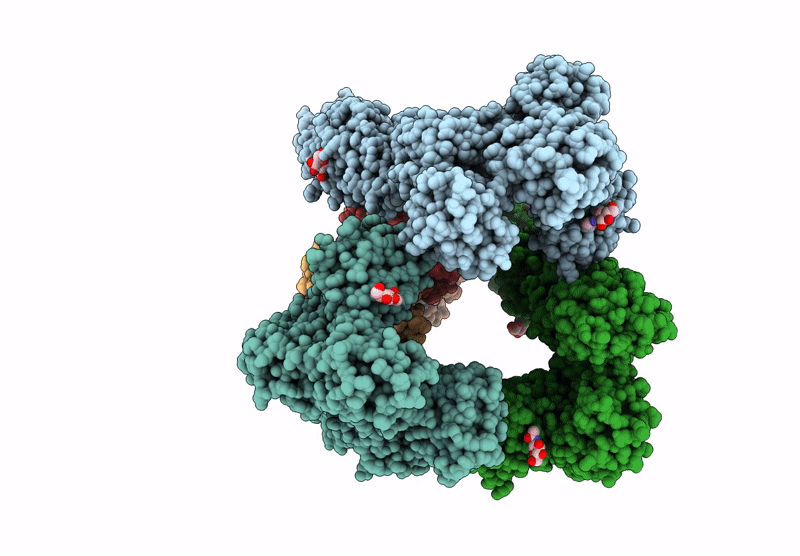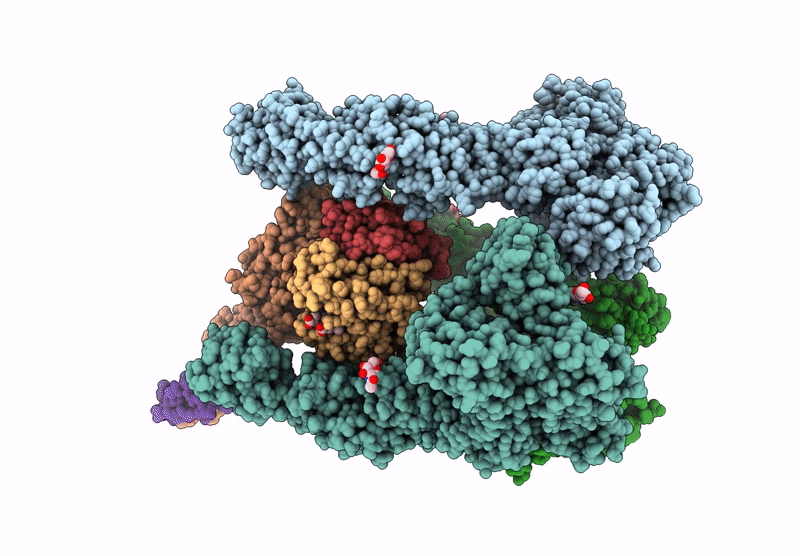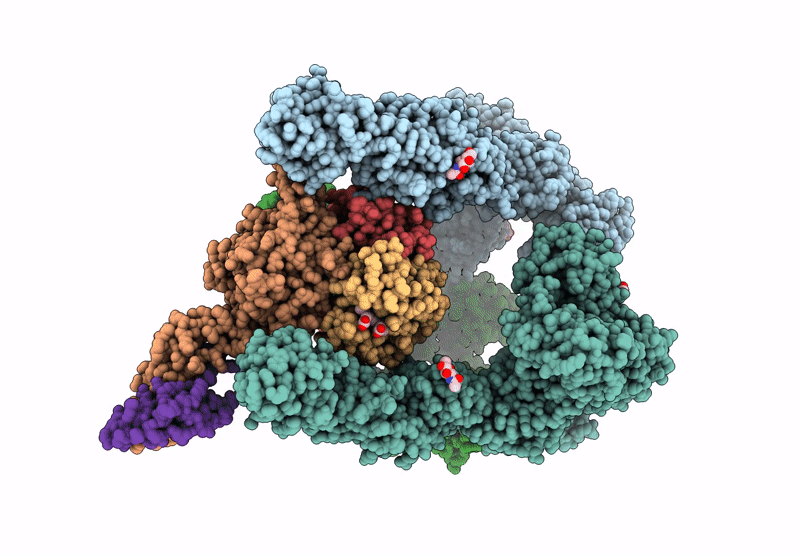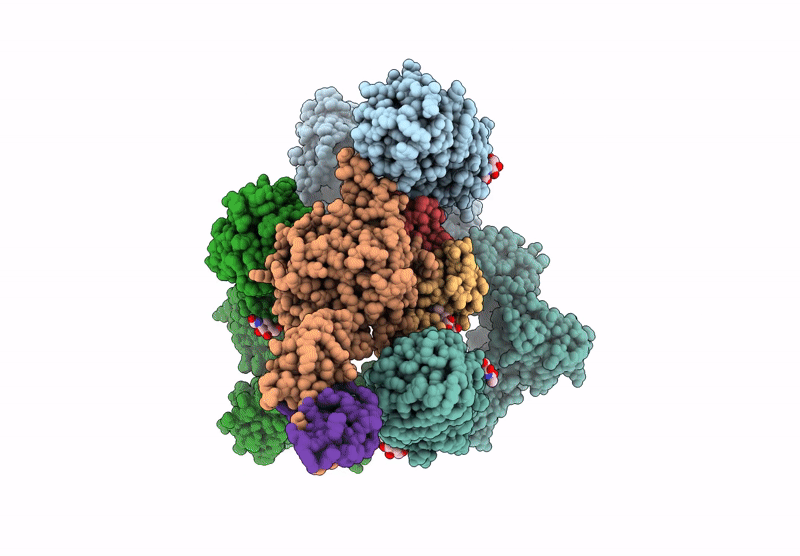
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ENDOCYTOSIS Ligands: CA, NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ENDOCYTOSIS Ligands: NAG, CA, HEM, OXY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ENDOCYTOSIS Ligands: CA, HEM, OXY |
 |
Organism: Bacillus methanolicus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-06-04 Classification: HYDROLASE |
 |
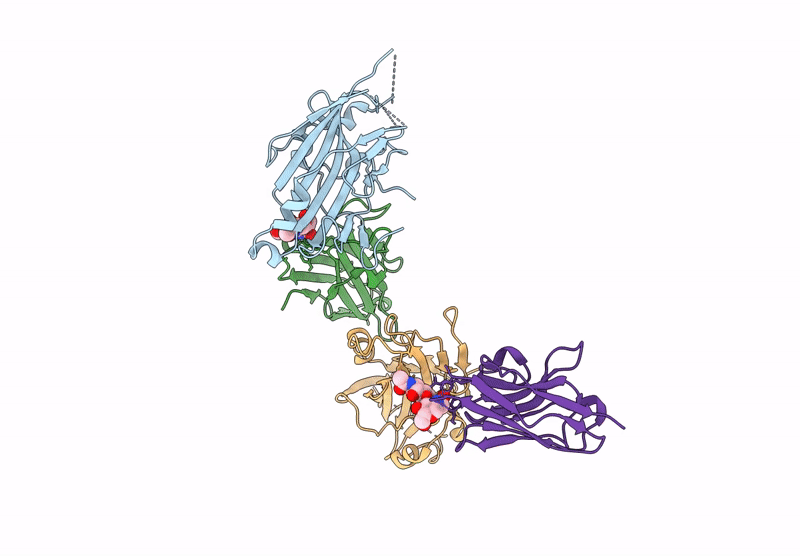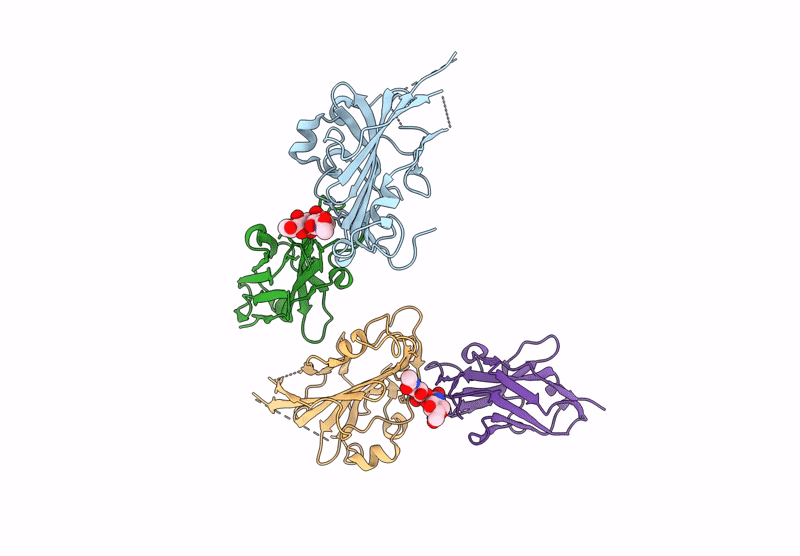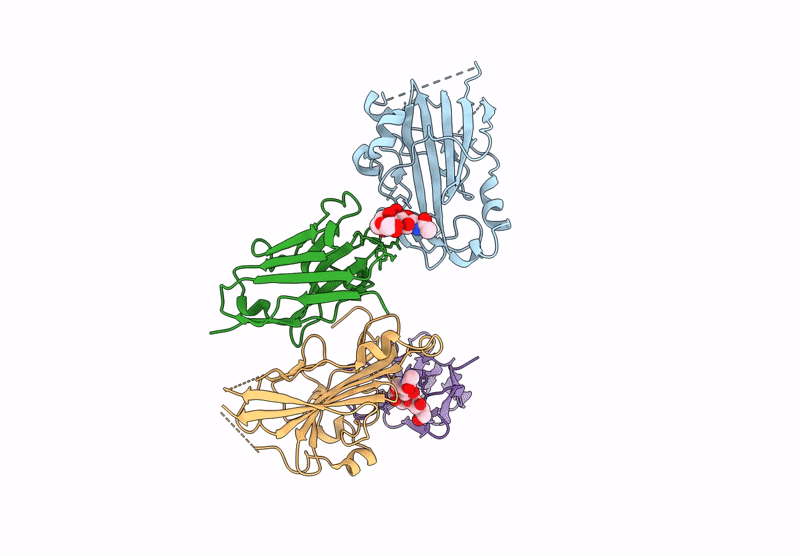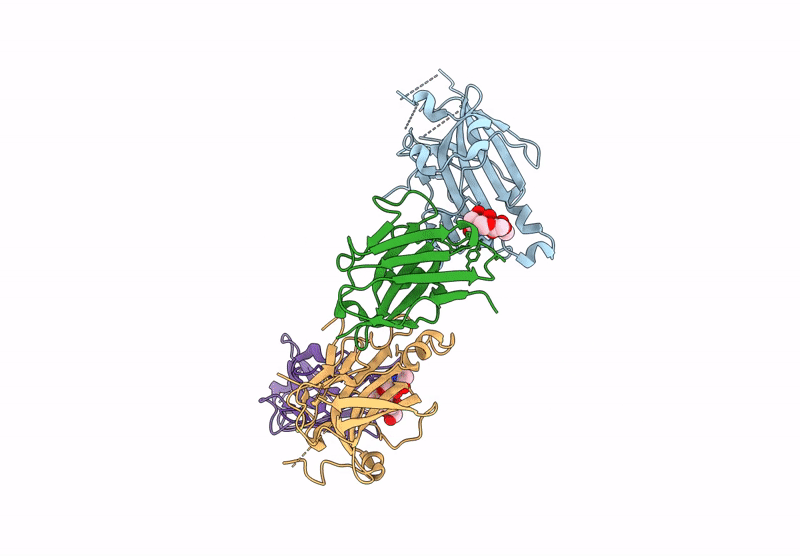
Organism: Human alphaherpesvirus 2, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:3.72 Å Release Date: 2025-05-28 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Human alphaherpesvirus 2, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2025-05-28 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |

