Search Count: 936
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PROTEIN BINDING Ligands: A1EHH |
 |
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Kirsten Rat Sarcoma G12C Complexed With Gmppnp And Covalently Bound To An Adduct Of {(2S)-4-[7-(8-Chloronaphthalen-1-Yl)-2-{[(2S)-1-Methylpyrrolidin-2-Yl]Methoxy}-5,6,7,8-Tetrahydropyrido[3,4-D]Pyrimidin-4-Yl]-1-[(2Z)-2-Fluoro-3-(Pyridin-2-Yl)Prop-2-Enoyl]Piperazin-2-Yl}Acetonitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GNP, A1B7P, MG |
 |
Crystal Structure Of Kirsten Rat Sarcoma G12C Complexed With Gdp And Covalently Bound To An Adduct Of (2S)-1-{4-[(7P)-7-(8-Ethynyl-7-Fluoro-3-Hydroxynaphthalen-1-Yl)-8-Fluoro-2-{[(2R,4R,7As)-2-Fluorotetrahydro-1H-Pyrrolizin-7A(5H)-Yl]Methoxy}Pyrido[4,3-D]Pyrimidin-4-Yl]Piperazin-1-Yl}-2-Fluoro-3-(1,3-Thiazol-2-Yl)Propan-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, A1B7Q, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
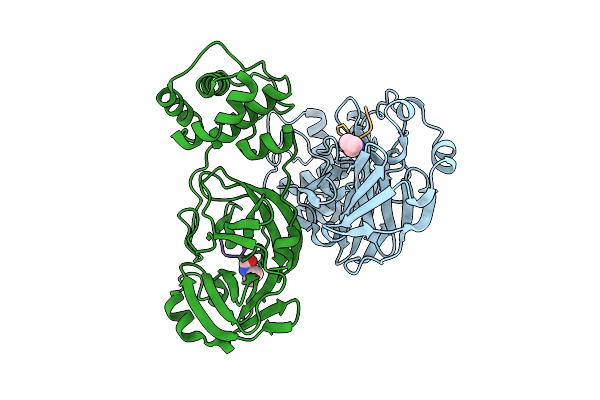 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBOTOR Ligands: A1L7M |
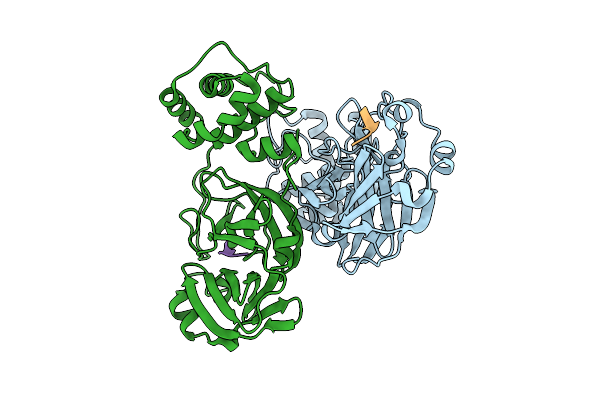 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
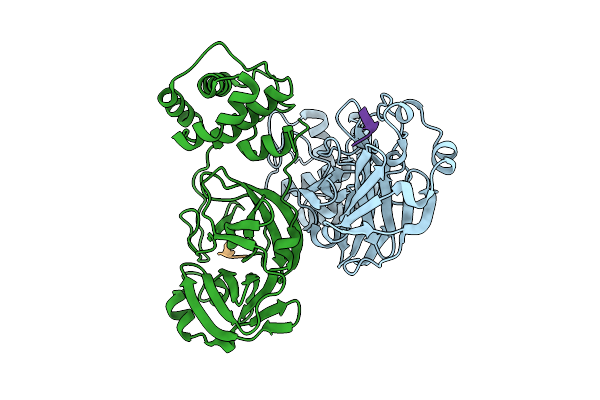 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
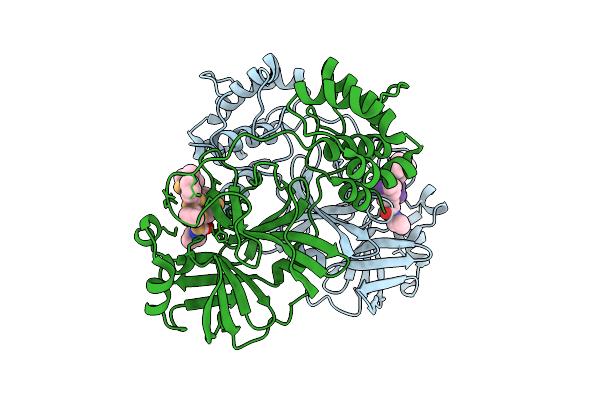 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN |
 |
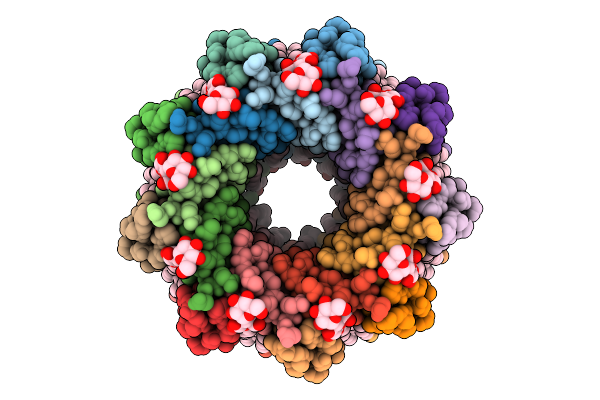
Light Harvesting Complex 3 (Lh3), B800-B820, Of Rhodoblastus (Rbl.) Acidophilus Strain 7750
Organism: Rhodoblastus acidophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PHOTOSYNTHESIS Ligands: BCL, LDA, IRM, LMT |
 |
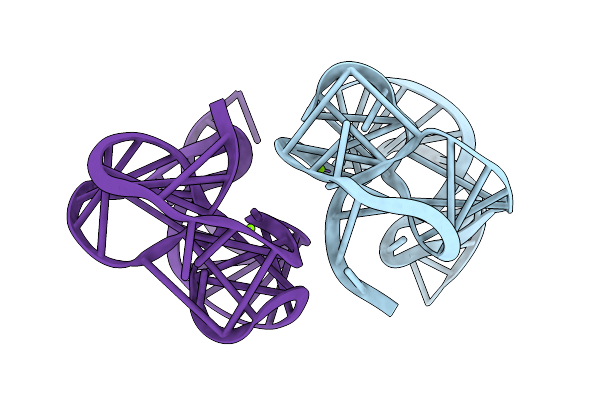
Light Harvesting Complex 2 (Lh2), B800-B850, Of Rhodoblastus (Rbl.) Acidophilus Strain 7750
Organism: Rhodoblastus acidophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PHOTOSYNTHESIS Ligands: BCL, LDA, LMT, IRM |
 |
Crystal Structure Of An Exoribonuclease-Resistant Rna From A Tombusvirus-Like Associated Rna
Organism: Beet western yellows st9 associated virus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: RNA Ligands: IRI, MG |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LIGASE Ligands: GFD, A1EMI |

