Search Count: 192
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: URC |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: R75 |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: A1AIJ |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: A1EAY |
 |
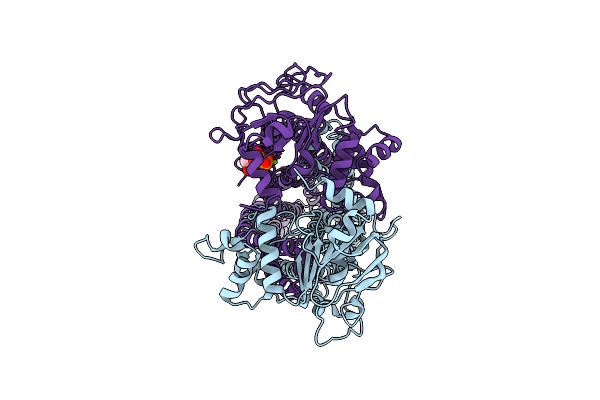
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp-Bound Ifasym-2 State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP |
 |
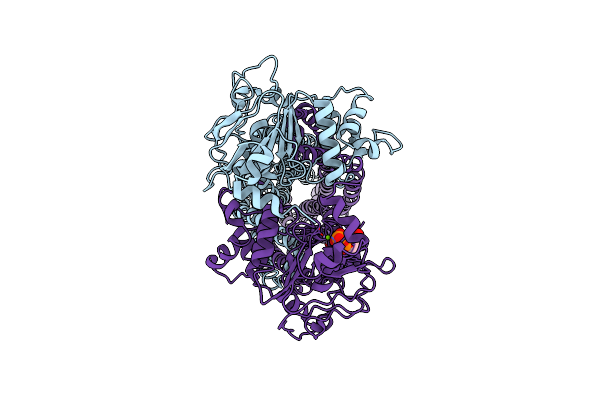
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Atp 37 Degrees C Treated
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
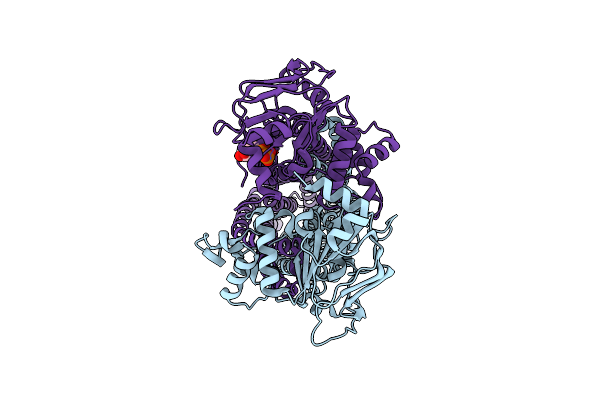
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Adp 4 Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
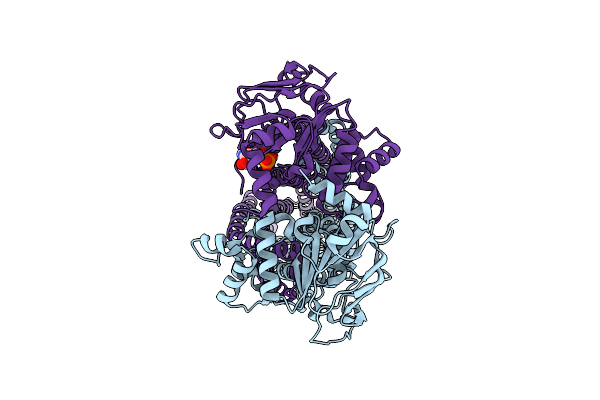
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 (Peptidisc) State (Atp 37Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 (Peptidisc) State (Adp 4Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp+Vi-Bound Occ (Vi) State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP, VO4 |
 |
Cryo-Em Structure Of Msrv1273C/72C(E553Q) Mutant From Mycobacterium Smegmatis In The Atp-Bound Occ State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Ifapo State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Amppnp-Bound Ifasym-1 State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: ANP, MG |
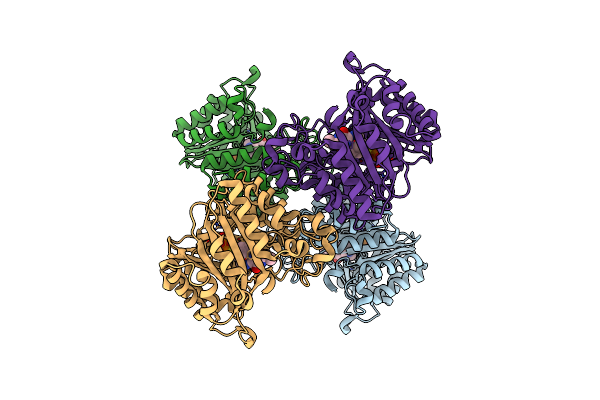 |
Versatile Aromatic Prenyltransferase Auraa In Complex With Dmapp And Cyclo-(L-Val-L-His)
Organism: Penicillium
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: DST, A1LYE |
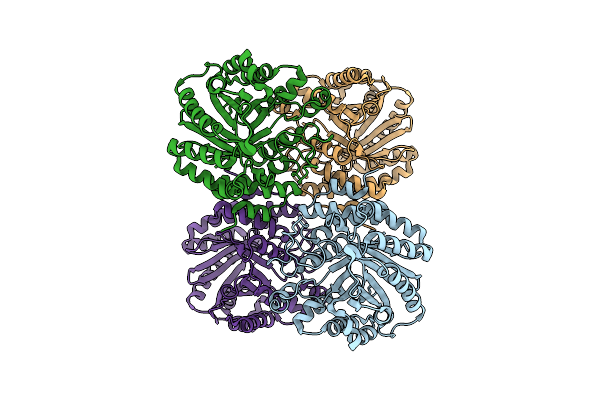 |
Versatile Aromatic Prenyltransferase Auraa For Imidazole-Containing Diketopiperazines
Organism: Penicillium
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE |
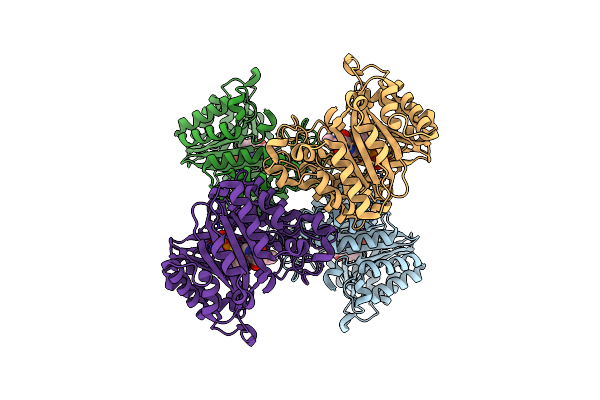 |
Versatile Aromatic Prenyltransferase Auraa In Complex With Dmspp And Cyclo-(L-Val-Dh-His)
Organism: Penicillium
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: DST, A1LYF |
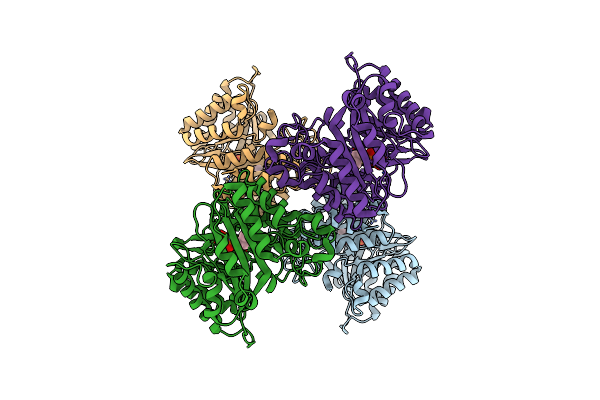 |
Versatile Aromatic Prenyltransferase Auraa Mutant-Y207A In Complex With Dmspp
Organism: Penicillium solitum
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: DST |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |

