Search Count: 20
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-12-11 Classification: OXYGEN TRANSPORT Ligands: GOL, HEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2024-12-11 Classification: OXYGEN TRANSPORT Ligands: HEM |
 |
Crystal Structure Of Arabidopsis Thaliana Acetohydroxyacid Synthase W574L Mutant In Complex With Chlorimuron-Ethyl
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-06-01 Classification: LIGASE Ligands: MG, FAD, CIE, TZD, NHE, SO4 |
 |
Crystal Structure Of Arabidopsis Thaliana Acetohydroxyacid Synthase P197T Mutant In Complex With Bispyribac-Sodium
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2022-06-01 Classification: LIGASE Ligands: MG, FAD, 6QL, AUJ, PO4, NHE, SO4 |
 |
Crystal Structure Of Arabidopsis Thaliana Acetohydroxyacid Synthase P197T Mutant In Complex With Chlorimuron-Ethyl
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2022-06-01 Classification: LIGASE Ligands: MG, FAD, CIE, NHE, F50, TP9, SO4 |
 |
Crystal Structure Of Arabidopsis Thaliana Acetohydroxyacid Synthase W574L Mutant
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2022-06-01 Classification: LIGASE Ligands: MG, FAD, TPP, NHE, SO4 |
 |
Crystal Structure Of Arabidopsis Thaliana Acetohydroxyacid Synthase W574L Mutant In Complex With Bispyribac-Sodium
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2022-06-01 Classification: LIGASE Ligands: MG, 6QL, FAD, TZD, NHE, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2012-10-17 Classification: SIGNALING PROTEIN |
 |
Organism: Viola arvensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-02-10 Classification: PLANT PROTEIN |
 |
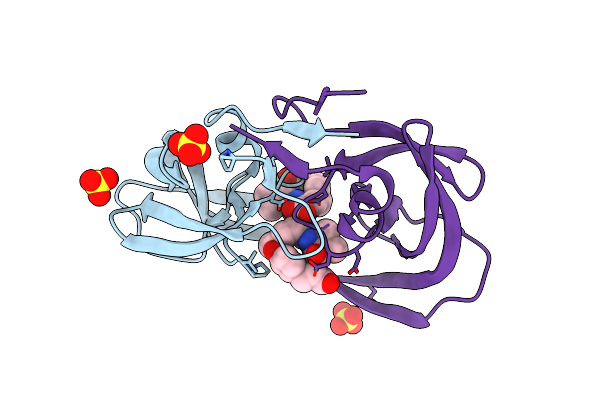
Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To Hiv-1 Protease. Implications For Catalytic Mechanism
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2008-03-25 Classification: HYDROLASE Ligands: SO4, DRR |
 |
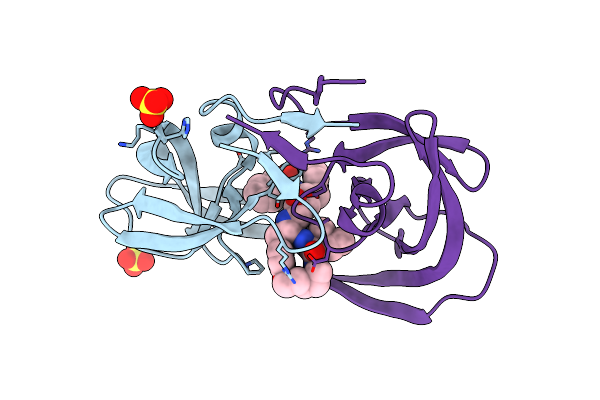
Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To Hiv-1 Protease. Implications For Catalytic Mechanism
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2008-03-25 Classification: HYDROLASE Ligands: SO4, DRS |
 |
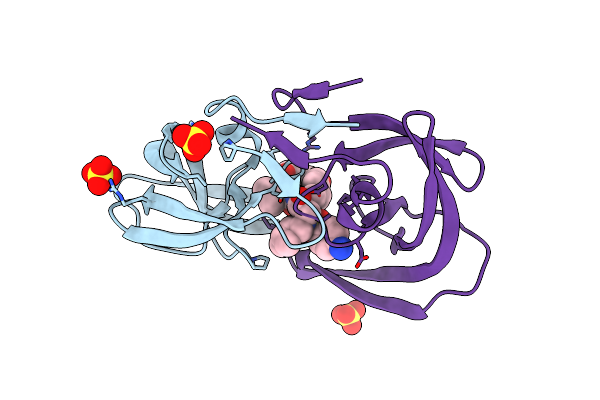
Method: X-RAY DIFFRACTION
Resolution:1.85 Å Release Date: 2000-10-11 Classification: HYDROLASE Ligands: SO4, PI8 |
 |
Method: X-RAY DIFFRACTION
Resolution:1.75 Å Release Date: 2000-10-11 Classification: HYDROLASE Ligands: SO4, PI9 |
 |
Organism: Conus episcopatus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 1999-01-13 Classification: ACETYLCHOLINE RECEPTOR ANTAGONIST |
 |
Organism: Conus pennaceus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 1998-05-20 Classification: ACETYLCHOLINE RECEPTOR ANTAGONIST |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1997-10-08 Classification: OXIDOREDUCTASE |
 |
Organism: Conus pennaceus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 1997-04-21 Classification: NEUROTOXIN |
 |
Structural Mechanism For Glycogen Phosphorylase Control By Phosphorylation And Amp
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1992-10-15 Classification: GLYCOGEN PHOSPHORYLASE Ligands: SO4, PLP |
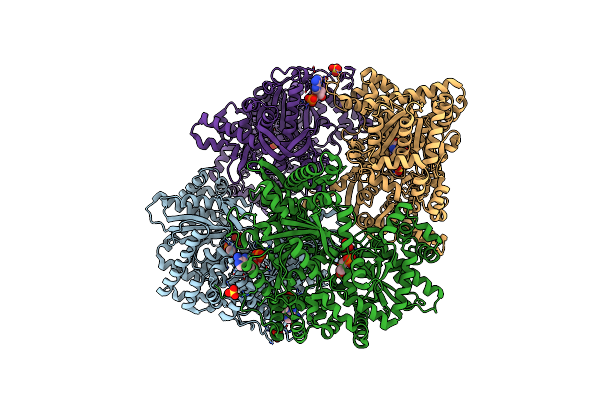 |
Structural Mechanism For Glycogen Phosphorylase Control By Phosphorylation And Amp
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1992-10-15 Classification: GLYCOGEN PHOSPHORYLASE Ligands: SO4, PLP, AMP |
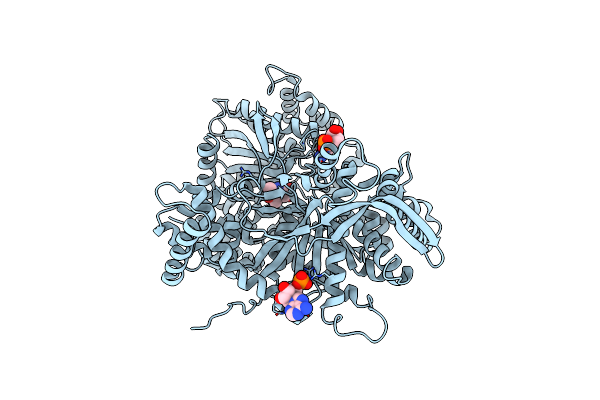 |
Structural Mechanism For Glycogen Phosphorylase Control By Phosphorylation And Amp
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1992-10-15 Classification: GLYCOGEN PHOSPHORYLASE Ligands: PLP, AMP |

